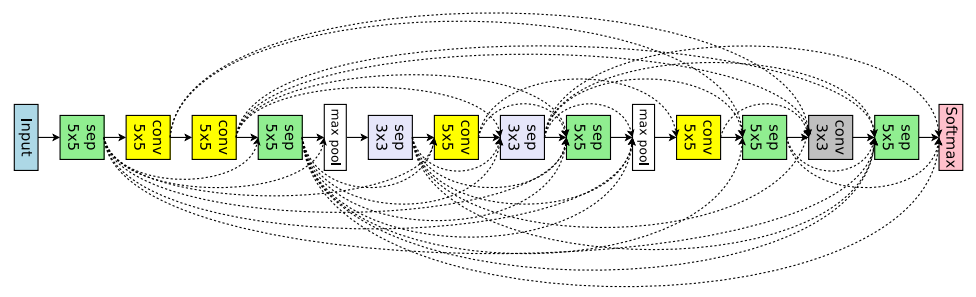
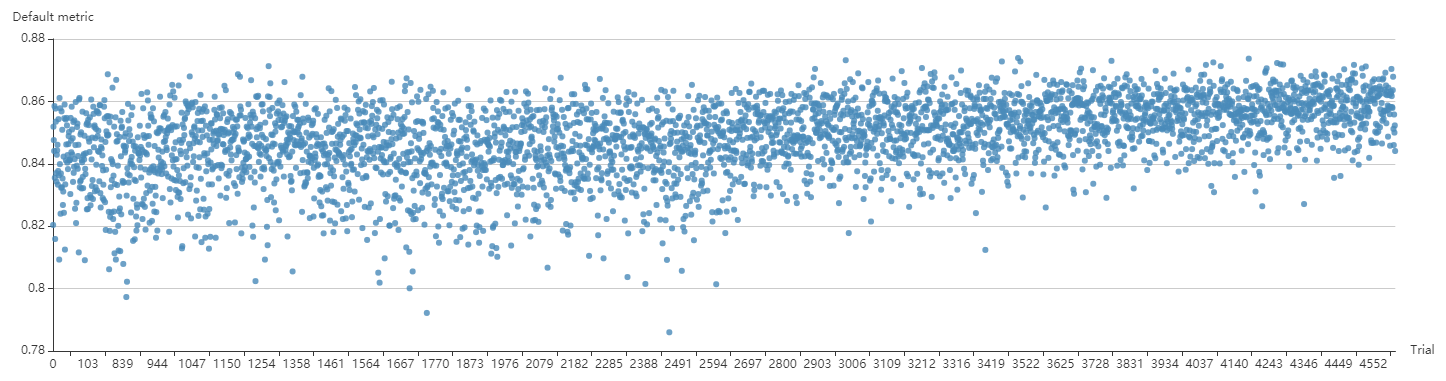
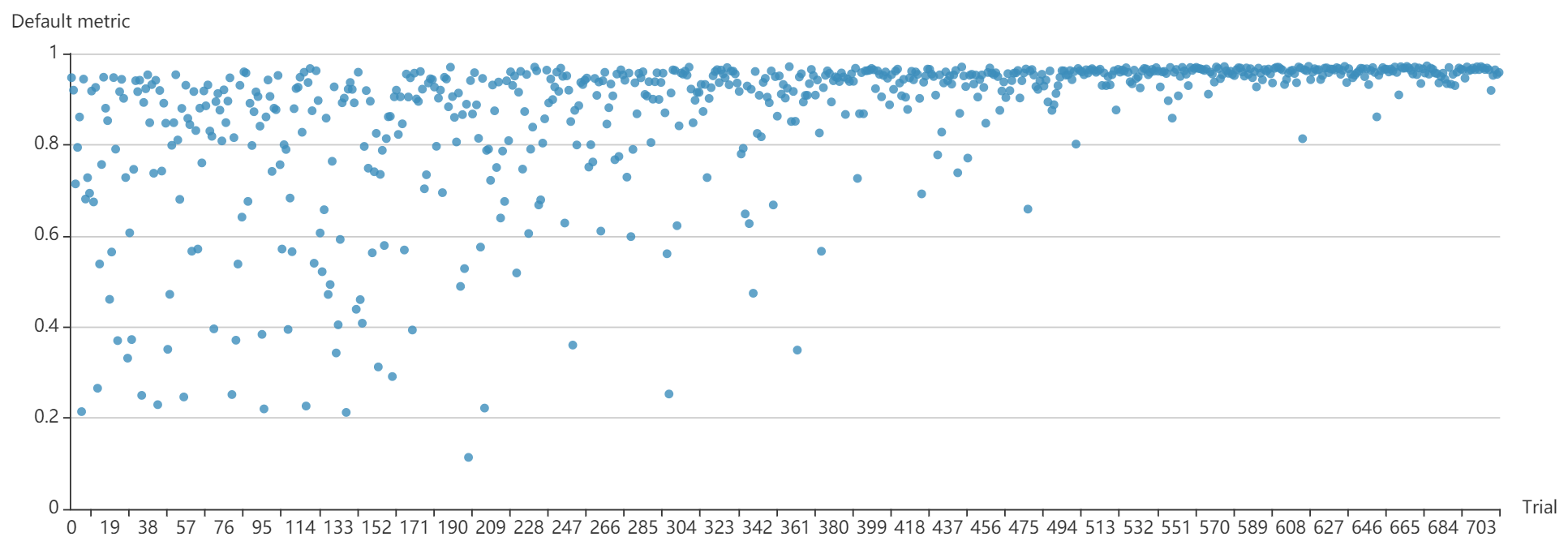
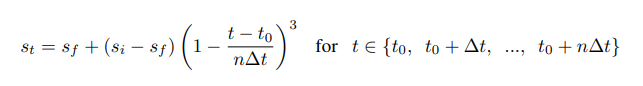"src/git@developer.sourcefind.cn:OpenDAS/nni.git" did not exist on "3fdbbdb3afbfe9564222a8e8a2381e1651a08a62"
Merge pull request #207 from microsoft/master
merge master
Showing
docs/en_US/Tuner/PPOTuner.md
0 → 100644
docs/img/agp_pruner.png
0 → 100644
8.38 KB
78.7 KB
docs/img/ppo_cifar10.png
0 → 100644
247 KB
docs/img/ppo_mnist.png
0 → 100644
99 KB