"src/lib/vscode:/vscode.git/clone" did not exist on "6c2cab25b1df103fe9aa08f2125c993a29fcf8cb"
Merge dev-nas-tuner back to master (#1531)
* PPO tuner for NAS, supports NNI's NAS interface (#1380)
Showing
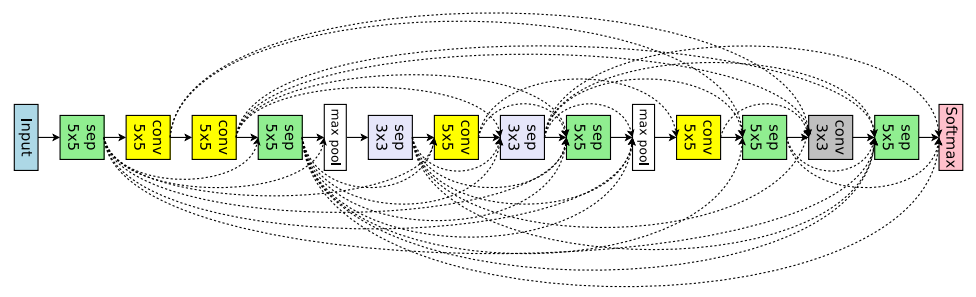
docs/en_US/Tuner/PPOTuner.md
0 → 100644
78.7 KB
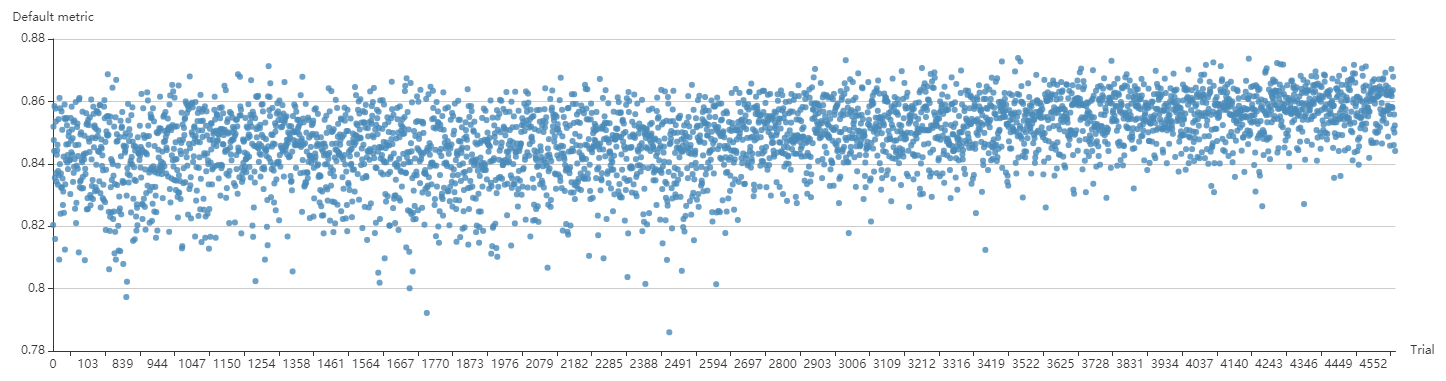
docs/img/ppo_cifar10.png
0 → 100644
247 KB
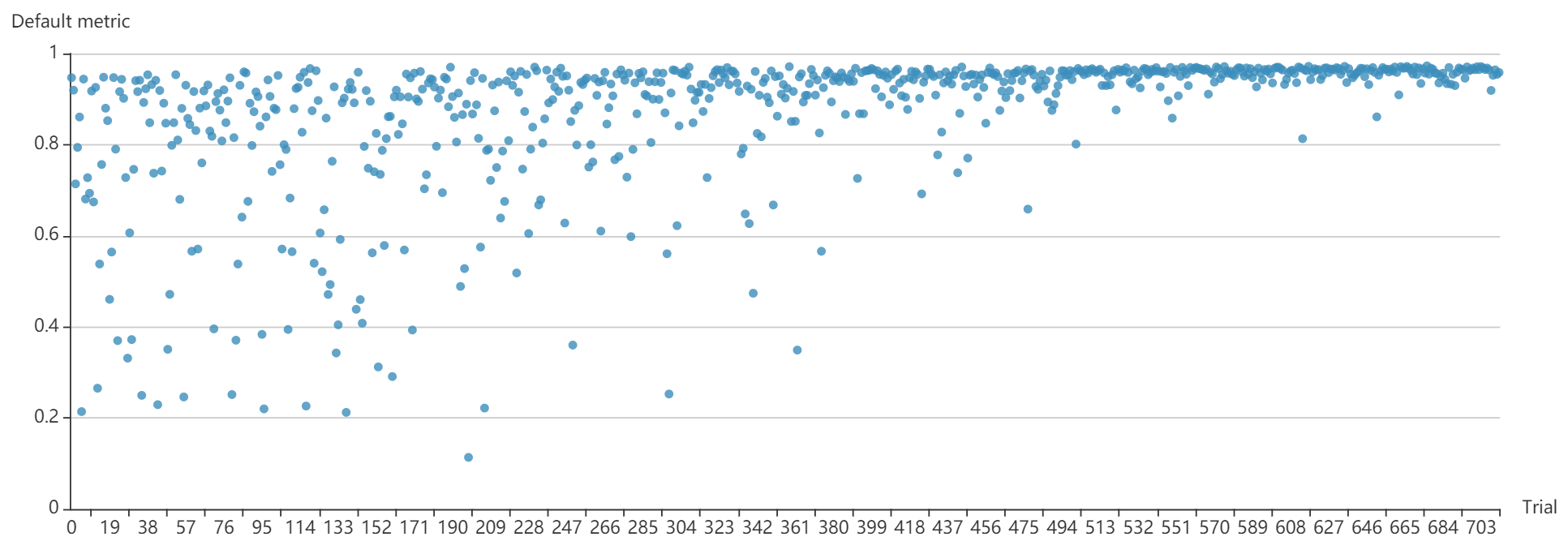
docs/img/ppo_mnist.png
0 → 100644
99 KB