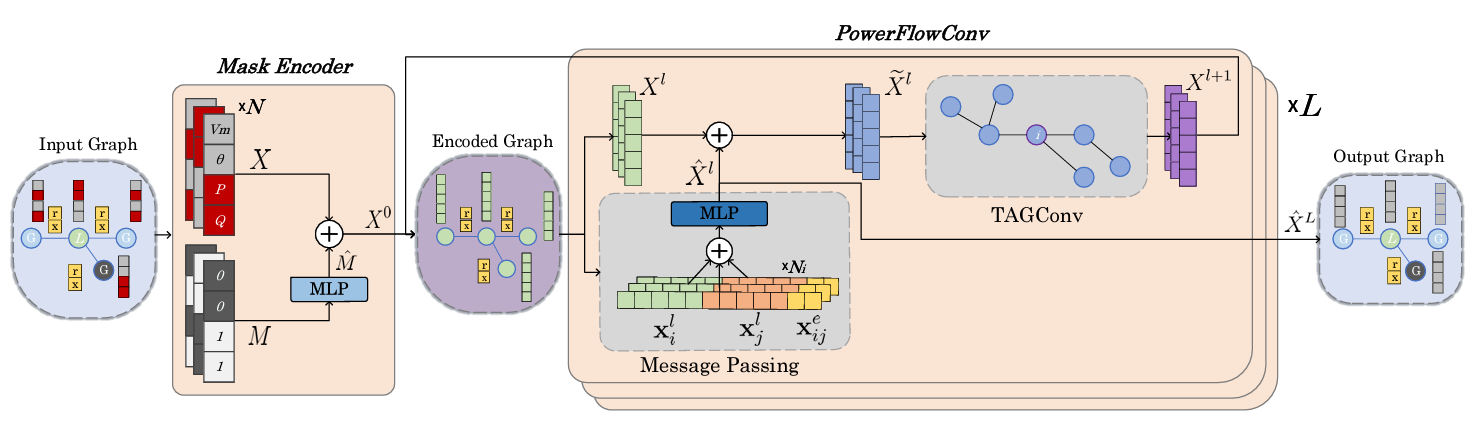
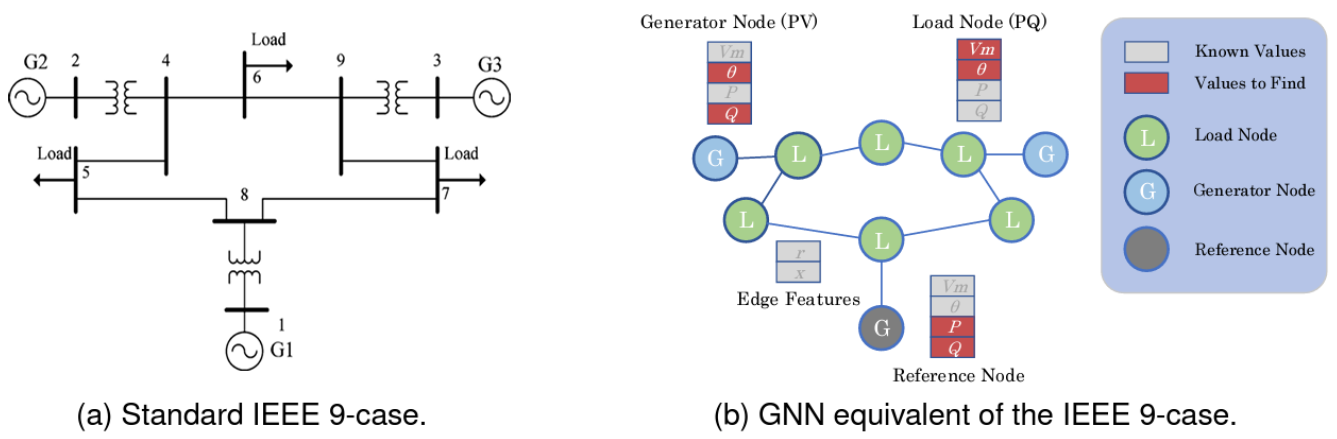
all
Showing
.gitignore
0 → 100644
LICENSE
0 → 100644
_loss_test.py
0 → 100644
collaborative_filtering.py
0 → 100644
configs/extra_large.json
0 → 100644
configs/large.json
0 → 100644
configs/small.json
0 → 100644
configs/standard.json
0 → 100644
dataset_generator.py
0 → 100644
dataset_generator_alt.py
0 → 100644
datasets/PowerFlowData.py
0 → 100644
datasets/__init__.py
0 → 100644
developing_transform.ipynb
0 → 100644
doc/arch_PF.png
0 → 100644
102 KB
doc/arch_PF1.png
0 → 100644
120 KB
docker/Dockerfile
0 → 100755
icon.png
0 → 100755
67.5 KB