容器环境下多体蛋白推理报错
环境及输入命令
环境:超算集群SothisAI容器
启动方式:参考光源docker pull,
实例化节点资源:单节点32核、100G、4*Z100
操作方式:
sudo su
cd /home
source env.sh
cd fastfold_pytorch
sh inference.py #此项通过
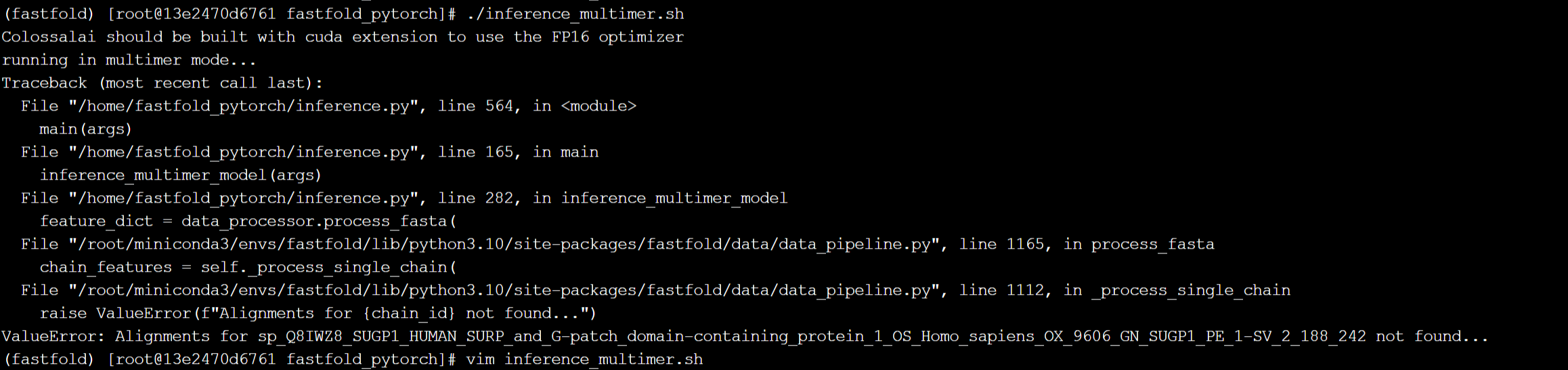
./inference_multimer.sh #此项报错蛋白质对齐报错
报错截图:
infer_multi脚本如下:
# add `--gpus [N]` to use N gpus for inference
# add `--enable_workflow` to use parallel workflow for data processing
# add `--use_precomputed_alignments [path_to_alignments]` to use precomputed msa
python3 inference.py SUGP1.fasta /public/home/jayfu/2024/data/pdb_mmcif/mmcif_files \
--output_dir ./ \
--gpus 1 \
--use_precomputed_alignments alignments/ \
--model_preset multimer \
--uniref90_database_path /public/home/jayfu/2024/data/uniref90/uniref90.fasta \
--mgnify_database_path /public/home/jayfu/2024/data/mgnify/mgy_clusters_2018_12.fa \
--pdb70_database_path /public/home/jayfu/2024/data/pdb70/pdb70 \
--uniclust30_database_path /public/home/jayfu/2024/data/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
--bfd_database_path /public/home/jayfu/2024/data/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--uniprot_database_path /public/home/jayfu/2024/data/uniprot/uniprot_sprot.fasta \
--pdb_seqres_database_path /public/home/jayfu/2024/data/pdb_seqres/pdb_seqres.txt \
--param_path /data/params/params_model_1_multimer.npz \
--model_name model_1_multimer \
--jackhmmer_binary_path `which jackhmmer` \
--hhblits_binary_path `which hhblits` \
--hhsearch_binary_path `which hhsearch` \
--kalign_binary_path `which kalign` \
--chunk_size 4 \
--inplace \推理报错
对比光源提供的示例命令,发现没有传入对齐蛋白参数,注释该行后出现新的错误:
Colossalai should be built with cuda extension to use the FP16 optimizer
running in monomer mode...
Colossalai should be built with cuda extension to use the FP16 optimizer
WARNING: Logging before InitGoogleLogging() is written to STDERR
I1204 14:44:37.992952 2030 ProcessGroupNCCL.cpp:686] [Rank 0] ProcessGroupNCCL initialization options:NCCL_ASYNC_ERROR_HANDLING: 1, NCCL_DESYNC_DEBUG: 0, NCCL_ENABLE_TIMING: 0, NCCL_BLOCKING_WAIT: 0, TIMEOUT(ms): 1800000, USE_HIGH_PRIORITY_STREAM: 0, TORCH_DISTRIBUTED_DEBUG: OFF, NCCL_DEBUG: OFF, ID=93946160889344
I1204 14:44:37.995162 2030 ProcessGroupNCCL.cpp:686] [Rank 0] ProcessGroupNCCL initialization options:NCCL_ASYNC_ERROR_HANDLING: 1, NCCL_DESYNC_DEBUG: 0, NCCL_ENABLE_TIMING: 0, NCCL_BLOCKING_WAIT: 0, TIMEOUT(ms): 1800000, USE_HIGH_PRIORITY_STREAM: 0, TORCH_DISTRIBUTED_DEBUG: OFF, NCCL_DEBUG: OFF, ID=93946160915856
I1204 14:44:37.995922 2030 ProcessGroupNCCL.cpp:686] [Rank 0] ProcessGroupNCCL initialization options:NCCL_ASYNC_ERROR_HANDLING: 1, NCCL_DESYNC_DEBUG: 0, NCCL_ENABLE_TIMING: 0, NCCL_BLOCKING_WAIT: 0, TIMEOUT(ms): 1800000, USE_HIGH_PRIORITY_STREAM: 0, TORCH_DISTRIBUTED_DEBUG: OFF, NCCL_DEBUG: OFF, ID=93946160928768
I1204 14:44:37.996516 2030 ProcessGroupNCCL.cpp:686] [Rank 0] ProcessGroupNCCL initialization options:NCCL_ASYNC_ERROR_HANDLING: 1, NCCL_DESYNC_DEBUG: 0, NCCL_ENABLE_TIMING: 0, NCCL_BLOCKING_WAIT: 0, TIMEOUT(ms): 1800000, USE_HIGH_PRIORITY_STREAM: 0, TORCH_DISTRIBUTED_DEBUG: OFF, NCCL_DEBUG: OFF, ID=93946160941696
[12/04/24 14:44:38] INFO colossalai - colossalai - INFO: /root/miniconda3/envs/fastfold/lib/python3.10/site-packages/colossalai/context/parallel_context.py:521 set_device
INFO colossalai - colossalai - INFO: process rank 0 is bound to device 0
I1204 14:44:38.882680 2030 ProcessGroupNCCL.cpp:1340] NCCL_DEBUG: N/A
INFO colossalai - colossalai - INFO: /root/miniconda3/envs/fastfold/lib/python3.10/site-packages/colossalai/context/parallel_context.py:557 set_seed
INFO colossalai - colossalai - INFO: initialized seed on rank 0, numpy: 1024, python random: 1024, ParallelMode.DATA: 1024, ParallelMode.TENSOR: 1024,the default parallel seed is ParallelMode.DATA.
INFO colossalai - colossalai - INFO: /root/miniconda3/envs/fastfold/lib/python3.10/site-packages/colossalai/initialize.py:117 launch
INFO colossalai - colossalai - INFO: Distributed environment is initialized, data parallel size: 1, pipeline parallel size: 1, tensor parallel size: 1
Traceback (most recent call last):
File "/home/fastfold_pytorch/inference.py", line 564, in <module>
main(args)
File "/home/fastfold_pytorch/inference.py", line 167, in main
inference_monomer_model(args)
File "/home/fastfold_pytorch/inference.py", line 448, in inference_monomer_model
torch.multiprocessing.spawn(inference_model, nprocs=args.gpus, args=(args.gpus, result_q, batch, args))
File "/usr/local/lib/python3.10/site-packages/torch/multiprocessing/spawn.py", line 246, in spawn
return start_processes(fn, args, nprocs, join, daemon, start_method="spawn")
File "/usr/local/lib/python3.10/site-packages/torch/multiprocessing/spawn.py", line 202, in start_processes
while not context.join():
File "/usr/local/lib/python3.10/site-packages/torch/multiprocessing/spawn.py", line 163, in join
raise ProcessRaisedException(msg, error_index, failed_process.pid)
torch.multiprocessing.spawn.ProcessRaisedException:
-- Process 0 terminated with the following error:
Traceback (most recent call last):
File "/usr/local/lib/python3.10/site-packages/torch/multiprocessing/spawn.py", line 74, in _wrap
fn(i, *args)
File "/home/fastfold_pytorch/inference.py", line 152, in inference_model
out = model(batch)
File "/usr/local/lib/python3.10/site-packages/torch/nn/modules/module.py", line 1518, in _wrapped_call_impl
return self._call_impl(*args, **kwargs)
File "/usr/local/lib/python3.10/site-packages/torch/nn/modules/module.py", line 1527, in _call_impl
return forward_call(*args, **kwargs)
File "/root/miniconda3/envs/fastfold/lib/python3.10/site-packages/fastfold/model/hub/alphafold.py", line 524, in forward
outputs, m_1_prev, z_prev, x_prev = self.iteration(
File "/root/miniconda3/envs/fastfold/lib/python3.10/site-packages/fastfold/model/hub/alphafold.py", line 236, in iteration
m_1_prev, z_prev = self.recycling_embedder(
File "/usr/local/lib/python3.10/site-packages/torch/nn/modules/module.py", line 1518, in _wrapped_call_impl
return self._call_impl(*args, **kwargs)
File "/usr/local/lib/python3.10/site-packages/torch/nn/modules/module.py", line 1527, in _call_impl
return forward_call(*args, **kwargs)
File "/root/miniconda3/envs/fastfold/lib/python3.10/site-packages/fastfold/model/fastnn/ops.py", line 1145, in forward
for i in range(0, para_dim, chunk_size):
UnboundLocalError: local variable 'para_dim' referenced before assignment
./inference_multimer.sh: line 9: --model_preset: command not found 请问有没有解决办法,以及我注意到代码仓库和镜像中均没有多体蛋白.npz参数,在哪里可以下载到