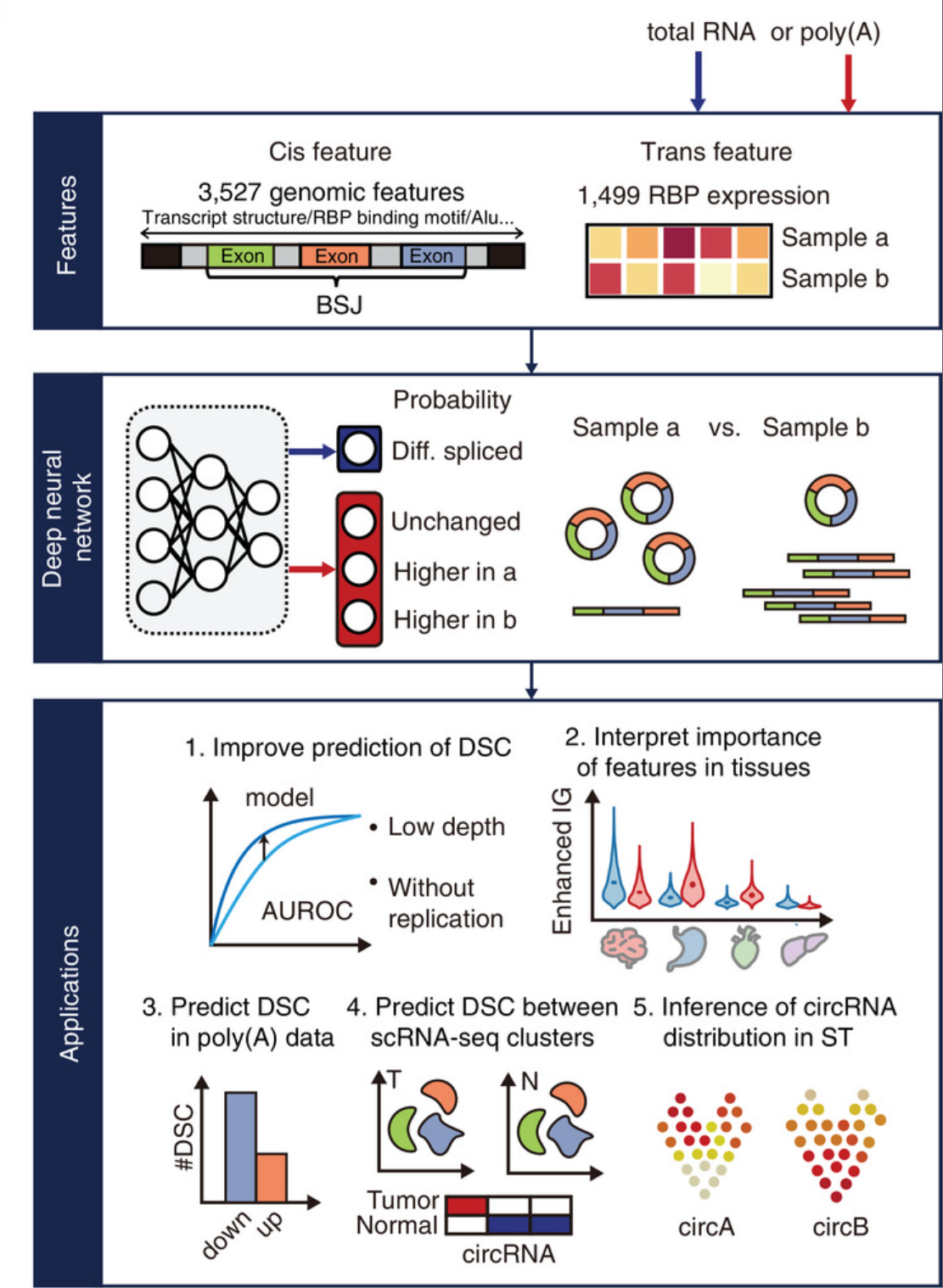
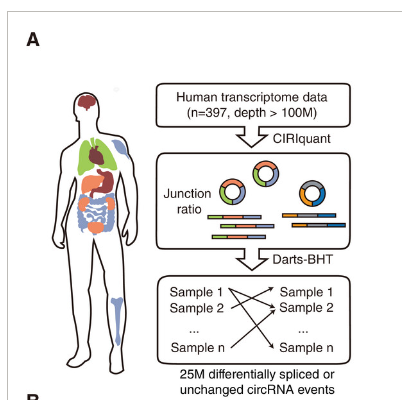
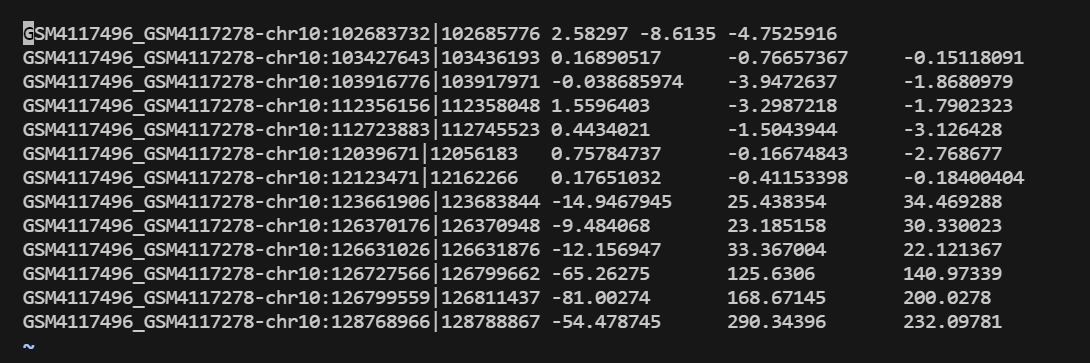
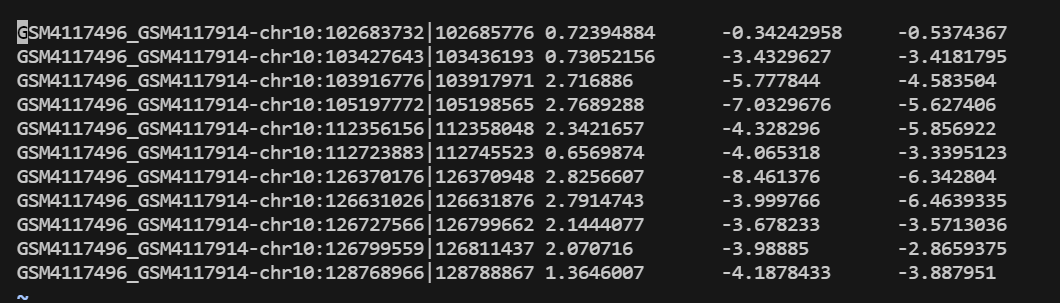
"examples/krr_classification_ex.cpp" did not exist on "1fc848fcf188cd88e1294047ddc63c93293da19b"
change readme.md
Showing
dockerfile
0 → 100644
images/image-1.png
0 → 100644
881 KB
images/image.png
0 → 100644
98.7 KB
images/image3.png
0 → 100644
75.2 KB
images/image4.png
0 → 100644
63.6 KB
| ... | @@ -3,4 +3,6 @@ matplotlib==3.3.4 | ... | @@ -3,4 +3,6 @@ matplotlib==3.3.4 |
| numpy==1.19.2 | numpy==1.19.2 | ||
| pandas==1.1.5 | pandas==1.1.5 | ||
| scikit-learn==0.24.2 | scikit-learn==0.24.2 | ||
| protobuf==3.20.1 | |||
| h5py<3.0.0 | |||
| #python | #python |