"magic_pdf/git@developer.sourcefind.cn:wangsen/mineru.git" did not exist on "eb02736a100794784168eebe80f25fe9b85aeda6"
Add documentation from 5.1
Showing
.nojekyll
0 → 100644
doc/html/.buildinfo
0 → 100644
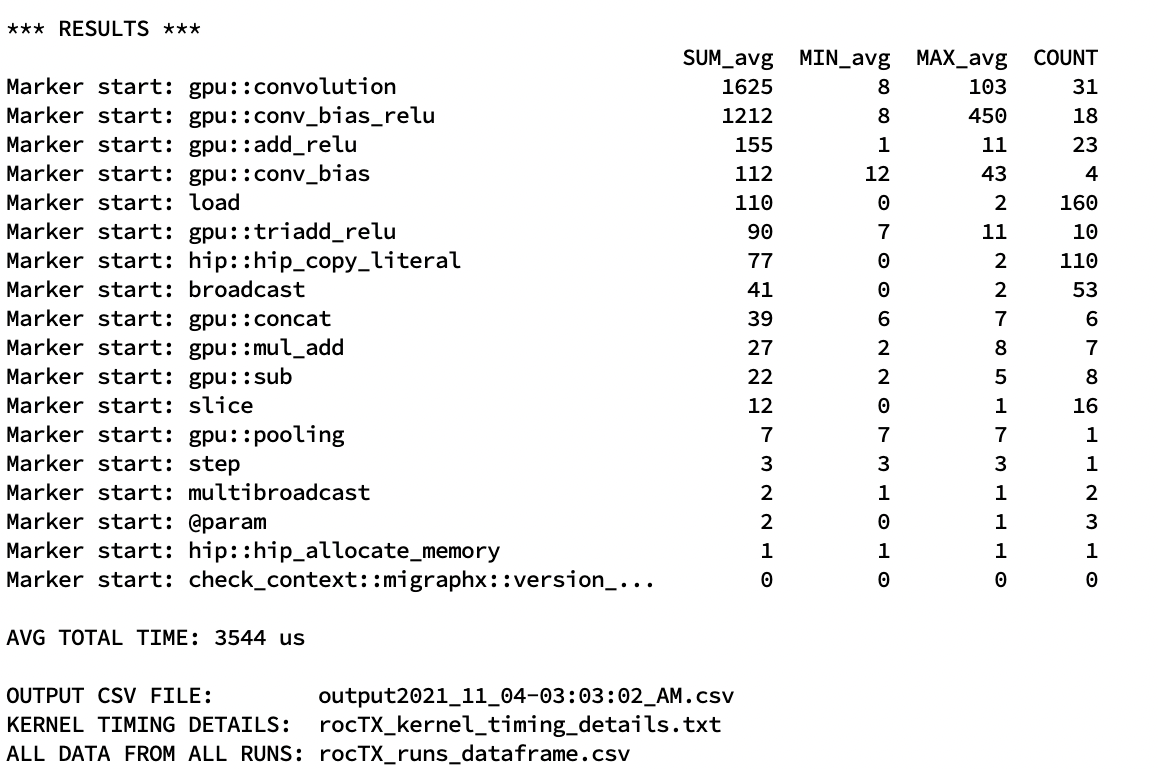
doc/html/_images/roctx1.jpg
0 → 100644
396 KB
doc/html/_images/roctx2.jpg
0 → 100644
123 KB