Merge branch 'develop' into mlir-c
Showing
doc/src/conf.py
100644 → 100755
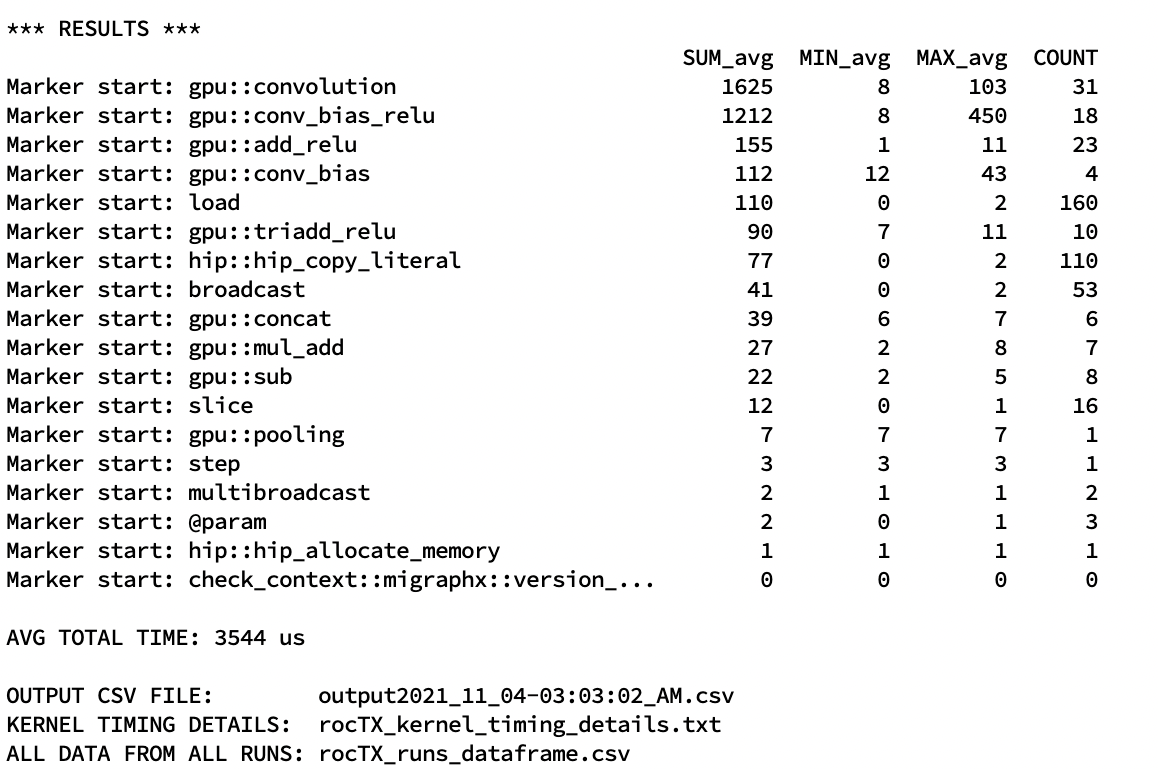
doc/src/dev/roctx1.jpg
0 → 100644
396 KB
doc/src/dev/roctx2.jpg
0 → 100644
123 KB
doc/src/dev/tools.rst
0 → 100644
src/targets/gpu/kernels/include/migraphx/kernels/functional.hpp
100755 → 100644