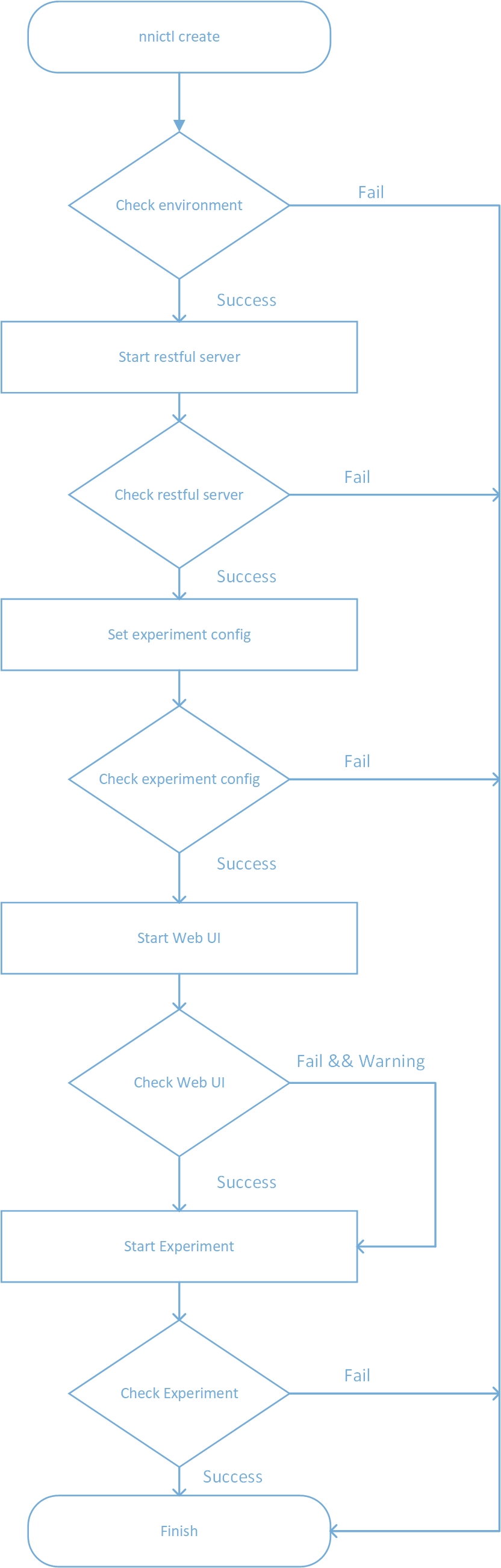
Refactor nnictl and add config_pai.yml (#144)
* fix nnictl bug * add hdfs host validation * fix bugs * fix dockerfile * fix install.sh * update install.sh * fix dockerfile * Set timeout for HDFSUtility exists function * remove unused TODO * fix sdk * add optional for outputDir and dataDir * refactor dockerfile.base * Remove unused import in hdfsclientUtility * add config_pai.yml * refactor nnictl create logic and add colorful print * fix nnictl stop logic * add annotation for config_pai.yml * add document for start experiment * fix config.yml * fix document
Showing
docs/StartExperiment.md
0 → 100644
296 KB