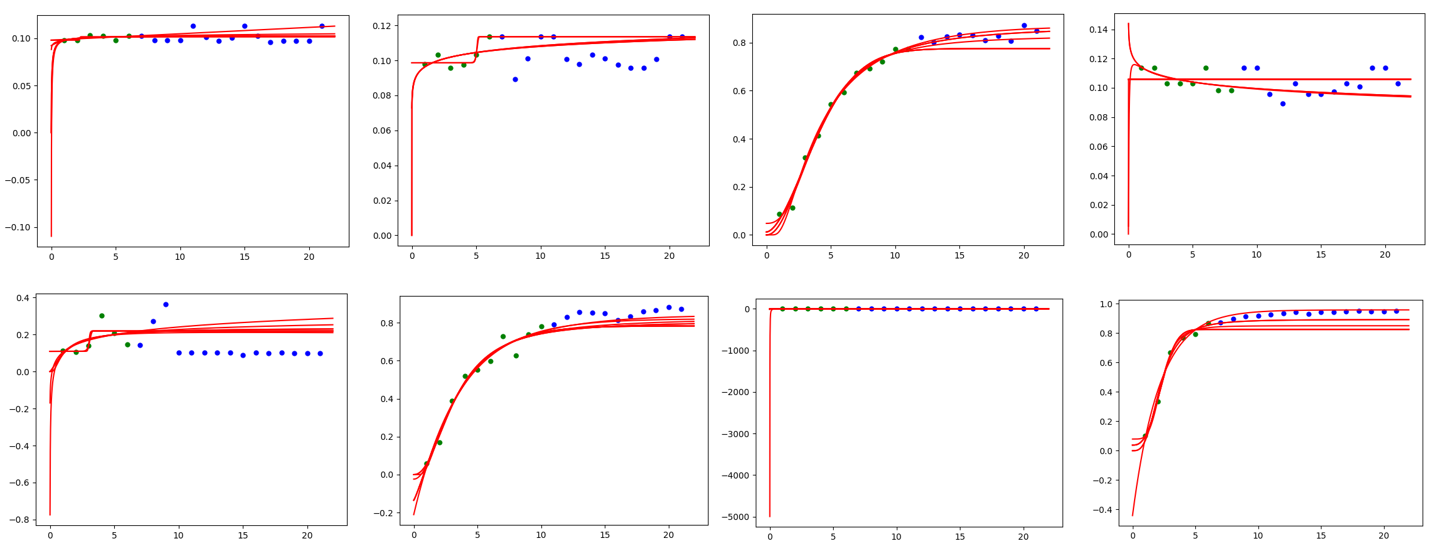
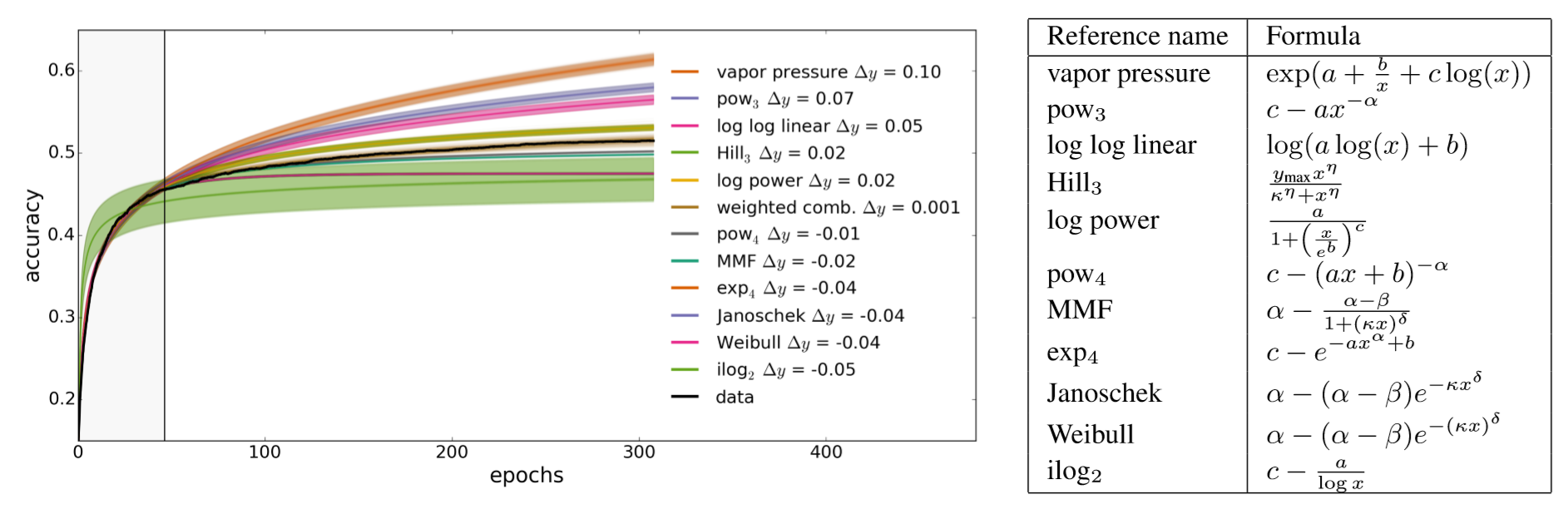
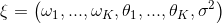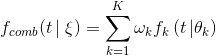"git@developer.sourcefind.cn:chenpangpang/diffusers.git" did not exist on "2c04e5855cf4a4696d8b1688e2858a93b125a865"
Merge pull request #135 from Microsoft/master
merge master
Showing
File moved
File moved
File moved
File moved
87.9 KB
docs/img/expression_xi.gif
0 → 100644
804 Bytes
docs/img/f_comb.gif
0 → 100644
1.17 KB
docs/img/learning_curve.PNG
0 → 100644
286 KB