"docs/git@developer.sourcefind.cn:change/sglang.git" did not exist on "48473684cc3e3d080fca85b089375700788f2d7a"
Merge pull request #2254 from microsoft/v1.5
Merge v1.5 branch back to master
Showing
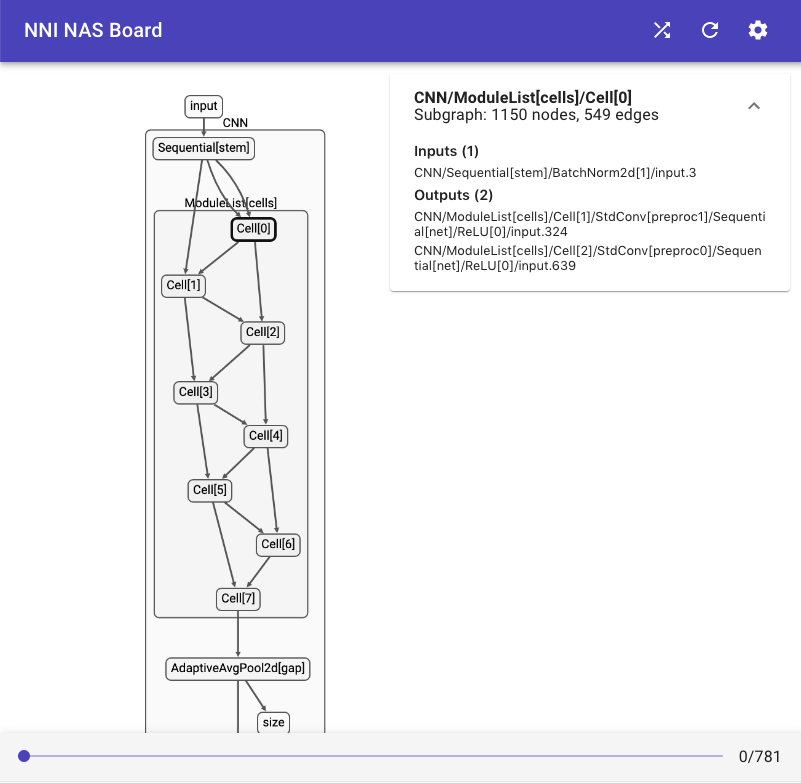
docs/img/nasui-1.png
0 → 100644
80.8 KB
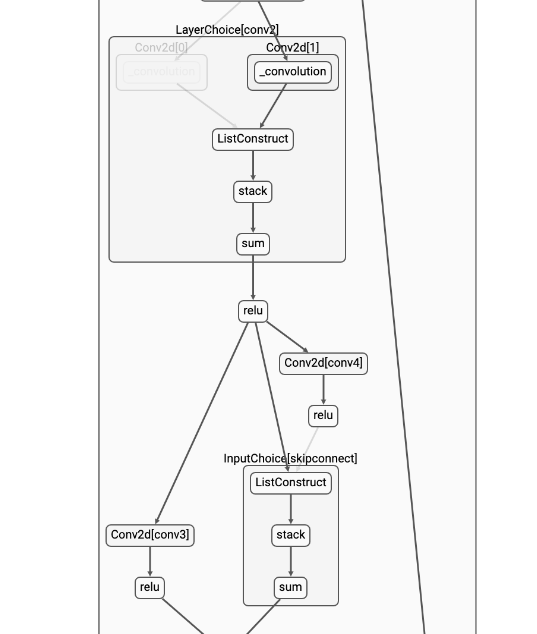
docs/img/nasui-2.png
0 → 100644
50.2 KB
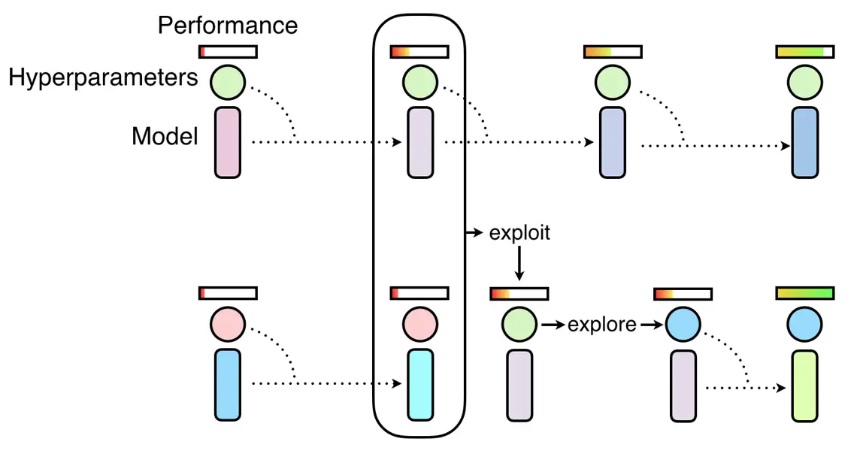
docs/img/pbt.jpg
0 → 100644
55 KB