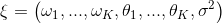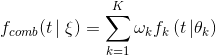"...resnet50_tensorflow.git" did not exist on "bd64315eb909fda4387b2a8e9e713bbbe184f6b9"
Add curve fitting assessor (#481)
* Add curve fitting assessor * Update HowToChooseTuner.md * Update HowToChooseTuner.md * Update HowToChooseTuner.md * Update README.md * Update README.md * Update README.md * Update HowToChooseTuner.md * Update HowToChooseTuner.md * Update HowToChooseTuner.md * Update HowToChooseTuner.md * Update curvefitting_assessor.py * Update config_schema.py * Add some comments and modifications * Remove unnecessary .json file * Remove unnecessary .lock file * Revert "Remove unnecessary .lock file" This reverts commit cdfaacb29114b3dee9c797d3e9b46ee18d7d34cc. * Revert "Revert "Remove unnecessary .lock file"" This reverts commit 7182a5fb31a02b01684429eabb3347952bf7ce2a. * Revert "Revert "Revert "Remove unnecessary .lock file""" This reverts commit 0f010e2b508e9f7b34c809647ba09e4e132876d8. * Revert "Remove unnecessary .json file" This reverts commit c6f7b47c199dd0db7ccb850d4f2ac1fd97b0caf8. * Revert "Add some comments and modifications" This reverts commit f78f055df9a4eec5b433a9241ce93d8ba78e3500. * Add some modifications by comments * suppoort minimize mode * Update README.md * Update README.md * Update modelfactory.py * minor changes and fix typo * minor chages * update README.md
Showing
87.9 KB
804 Bytes
1.17 KB