"docs/vscode:/vscode.git/clone" did not exist on "78b87dc25aa3cb5eab282354d9b001b90a75cca4"
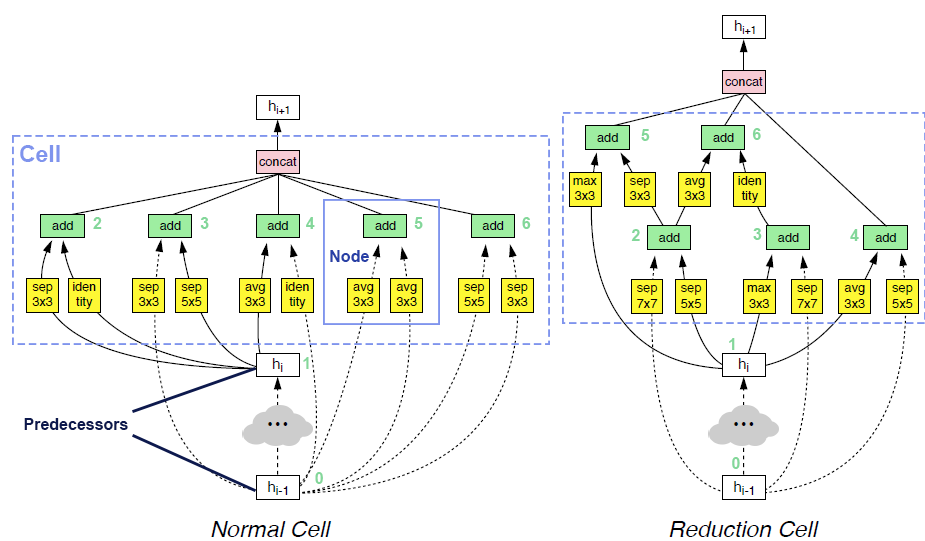
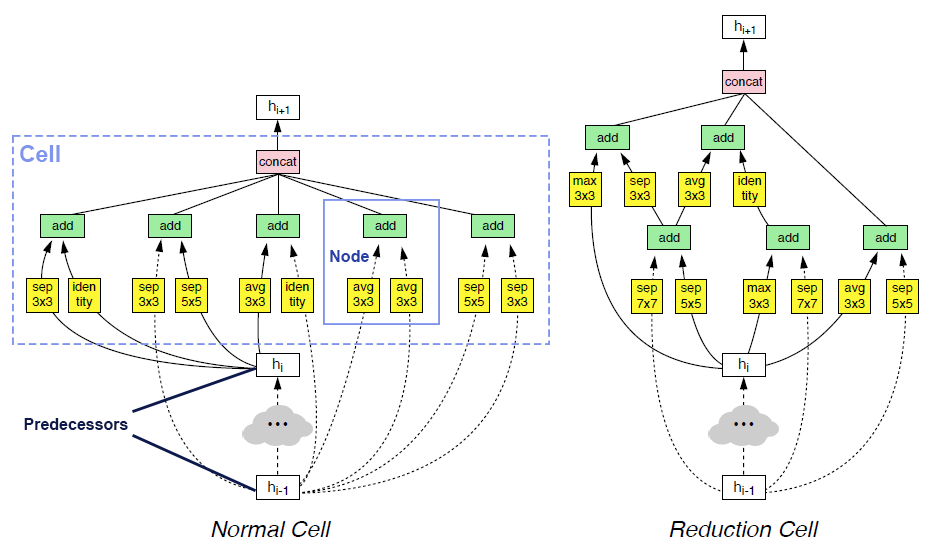
More improvements on NAS documentation and cell interface (#4752)
Showing
| W: | H:
| W: | H:


docs/source/nas/toctree.rst
0 → 100644
No preview for this file type