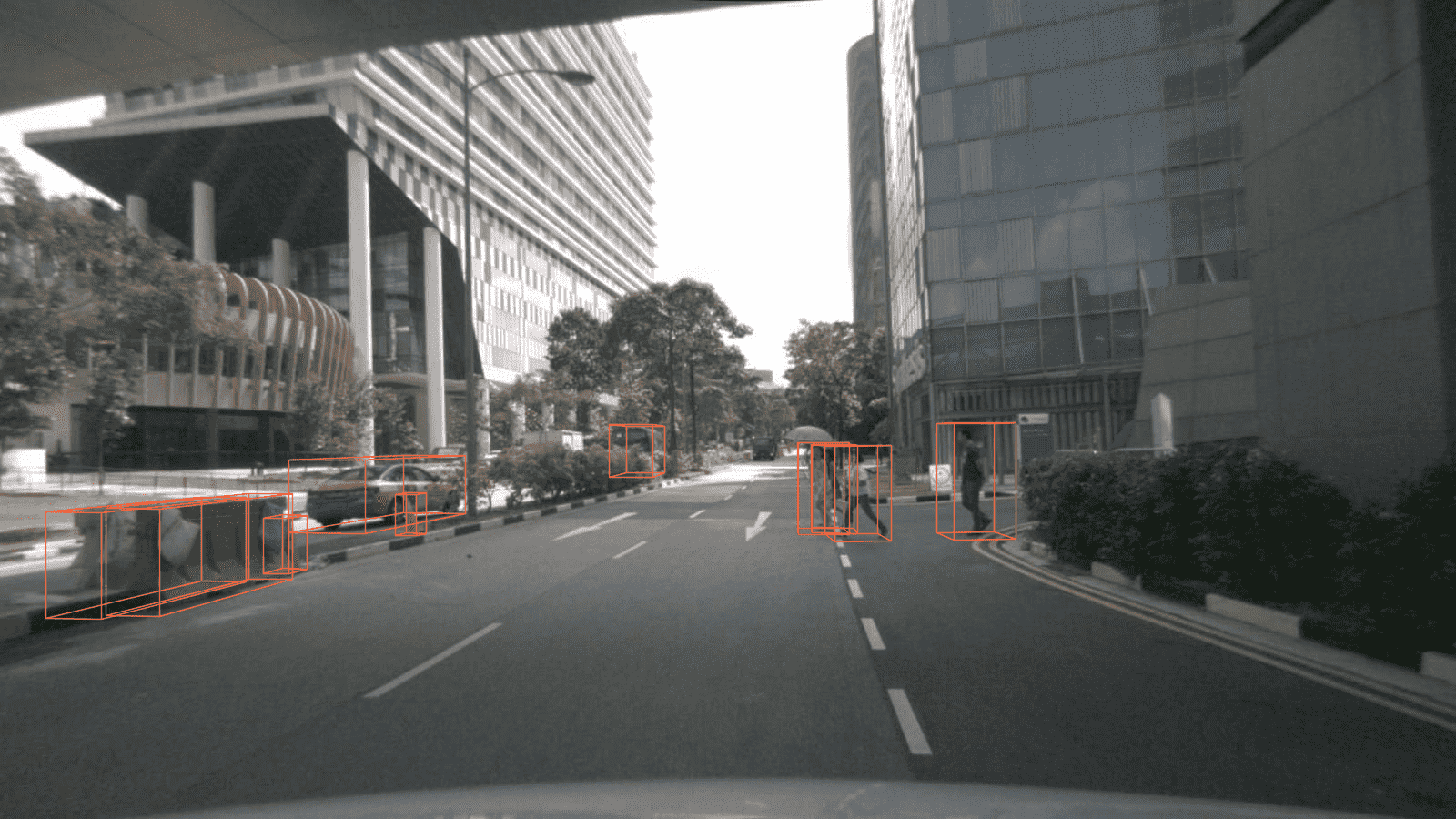
[Feature] Support visualization and browse_dataset on nuScenes Mono-3D dataset (#542)
* fix comment errors * add eval_pipeline in mono-nuscene cfg * add vis function in nuscene-mono dataset * refactor vis function to support all three mode boxes proj to img * add unit test for nuscene-mono show func * browse_dataset support nuScenes_mono * add show_results() to SingleStageMono3DDetector * support mono-3d dataset browse * support nus_mono_dataset and single_stage_mono_detector show function * update useful_tools.md docs * support mono-3d demo * add unit test * update demo docs * fix typos & remove unused comments * polish docs
Showing
141 KB
demo/mono_det_demo.py
0 → 100644
328 KB

| W: | H:
| W: | H: