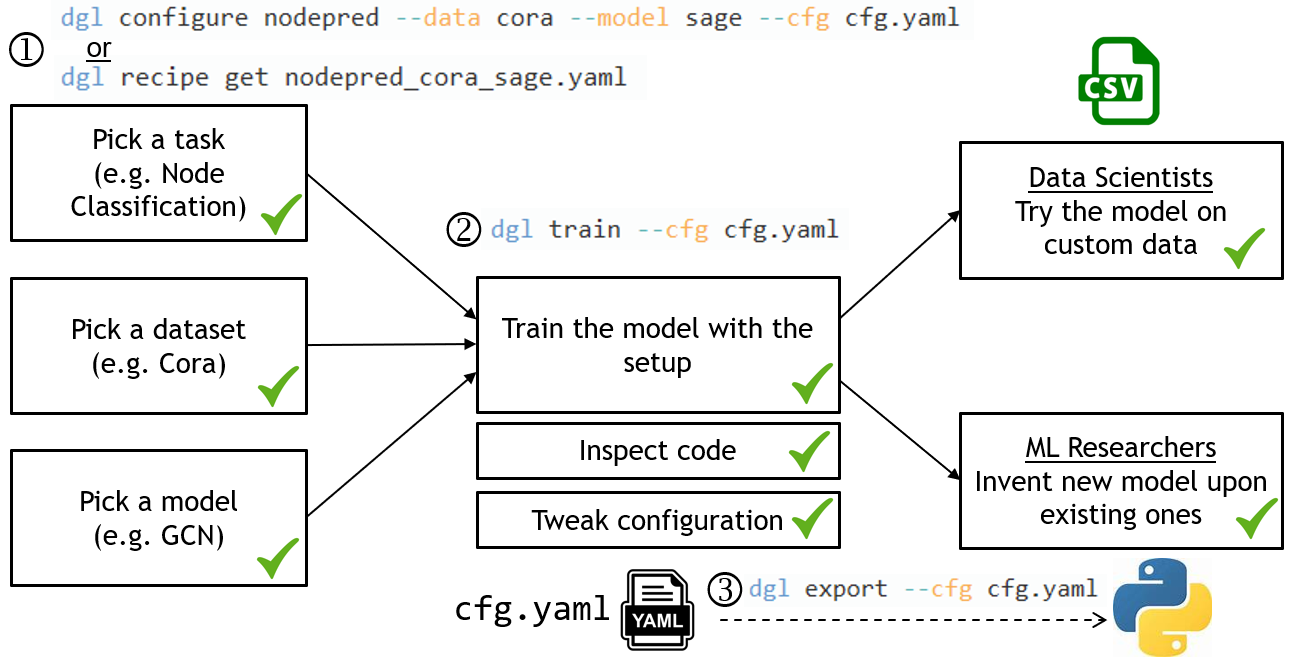
[DGL-Go] Change name to dglgo (#3778)
* add * remove * fix * rework the readme and some changes * add png * update png * add recipe get Co-authored-by:Minjie Wang <wmjlyjemaine@gmail.com> Co-authored-by:
Quan (Andy) Gan <coin2028@hotmail.com>
Showing
dglgo/dglgo.png
0 → 100644
147 KB
File moved
File moved
File moved
File moved
File moved
File moved