# U-KAN Makes Strong Backbone for Medical Image Segmentation and Generation
:pushpin: This is an official PyTorch implementation of **U-KAN Makes Strong Backbone for Medical Image Segmentation and Generation**
[[`Project Page`](https://yes-ukan.github.io/)] [[`arXiv`](https://arxiv.org/abs/2406.02918)] [[`BibTeX`](#citation)]

> [**U-KAN Makes Strong Backbone for Medical Image Segmentation and Generation**](https://arxiv.org/abs/2406.02918)
> [Chenxin Li](https://xggnet.github.io/)\*, [Xinyu Liu](https://xinyuliu-jeffrey.github.io/)\*, [Wuyang Li](https://wymancv.github.io/wuyang.github.io/)\*, [Cheng Wang](https://scholar.google.com/citations?user=AM7gvyUAAAAJ&hl=en)\*, [Hengyu Liu](), [Yixuan Yuan](https://www.ee.cuhk.edu.hk/~yxyuan/people/people.htm)✉
The Chinese Univerisity of Hong Kong
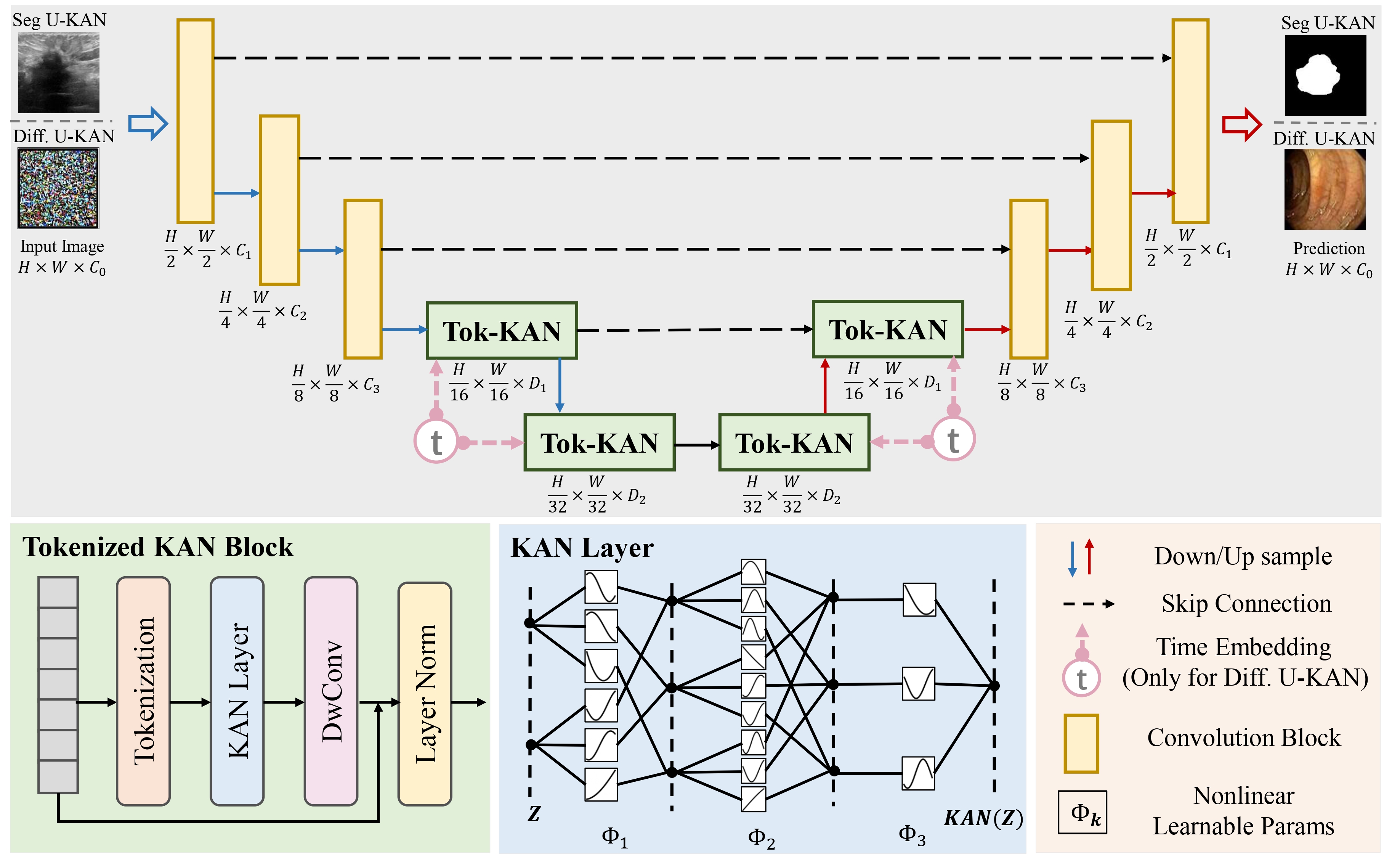
We explore the untapped potential of Kolmogorov-Anold Network (aka. KAN) in improving backbones for vision tasks. We investigate, modify and re-design the established U-Net pipeline by integrating the dedicated KAN layers on the tokenized intermediate representation, termed U-KAN. Rigorous medical image segmentation benchmarks verify the superiority of U-KAN by higher accuracy even with less computation cost. We further delved into the potential of U-KAN as an alternative U-Net noise predictor in diffusion models, demonstrating its applicability in generating task-oriented model architectures. These endeavours unveil valuable insights and sheds light on the prospect that with U-KAN, you can make strong backbone for medical image segmentation and generation.
## 📰News
**[2024.6]** Some modifications are done in Seg_UKAN for better performance reproduction. The previous code can be quickly updated by replacing the contents of train.py and archs.py with the new ones.
**[2024.6]** Model checkpoints and training logs are released!
**[2024.6]** Code and paper of U-KAN are released!
## 💡Key Features
- The first effort to incorporate the advantage of emerging KAN to improve established U-Net pipeline to be more **accurate, efficient and interpretable**.
- A Segmentation U-KAN with **tokenized KAN block to effectively steer the KAN operators** to be compatible with the exiting convolution-based designs.
- A Diffusion U-KAN as an **improved noise predictor** demonstrates its potential in backboning generative tasks and broader vision settings.
## 🛠Setup
```bash
git clone https://github.com/CUHK-AIM-Group/U-KAN.git
cd U-KAN
conda create -n ukan python=3.10
conda activate ukan
cd Seg_UKAN && pip install -r requirements.txt
```
**Tips A**: We test the framework using pytorch=1.13.0, and the CUDA compile version=11.6. Other versions should be also fine but not totally ensured.
## 📚Data Preparation
**BUSI**: The dataset can be found [here](https://www.kaggle.com/datasets/aryashah2k/breast-ultrasound-images-dataset).
**GLAS**: The dataset can be found [here](https://websignon.warwick.ac.uk/origin/slogin?shire=https%3A%2F%2Fwarwick.ac.uk%2Fsitebuilder2%2Fshire-read&providerId=urn%3Awarwick.ac.uk%3Asitebuilder2%3Aread%3Aservice&target=https%3A%2F%2Fwarwick.ac.uk%2Ffac%2Fcross_fac%2Ftia%2Fdata%2Fglascontest&status=notloggedin).
**CVC-ClinicDB**: The dataset can be found [here](https://www.dropbox.com/s/p5qe9eotetjnbmq/CVC-ClinicDB.rar?e=3&dl=0).
We also provide all the [pre-processed dataset](https://mycuhk-my.sharepoint.com/:f:/g/personal/1155206760_link_cuhk_edu_hk/ErDlT-t0WoBNlKhBlbYfReYB-iviSCmkNRb1GqZ90oYjJA?e=hrPNWD) without requiring any further data processing. You can directly download and put them into the data dir.
The resulted file structure is as follows.
```
Seg_UKAN
├── inputs
│ ├── busi
│ ├── images
│ ├── malignant (1).png
| ├── ...
| ├── masks
│ ├── 0
│ ├── malignant (1)_mask.png
| ├── ...
│ ├── GLAS
│ ├── images
│ ├── 0.png
| ├── ...
| ├── masks
│ ├── 0
│ ├── 0.png
| ├── ...
│ ├── CVC-ClinicDB
│ ├── images
│ ├── 0.png
| ├── ...
| ├── masks
│ ├── 0
│ ├── 0.png
| ├── ...
```
## 🔖Evaluating Segmentation U-KAN
You can directly evaluate U-KAN from the checkpoint model. Here is an example for quick usage for using our **pre-trained models** in [Segmentation Model Zoo](#segmentation-model-zoo):
1. Download the pre-trained weights and put them to ```{args.output_dir}/{args.name}/model.pth```
2. Run the following scripts to
```bash
cd Seg_UKAN
python val.py --name ${dataset}_UKAN --output_dir [YOUR_OUTPUT_DIR]
```
## ⏳Training Segmentation U-KAN
You can simply train U-KAN on a single GPU by specifing the dataset name ```--dataset``` and input size ```--input_size```.
```bash
cd Seg_UKAN
python train.py --arch UKAN --dataset ${dataset} --input_w ${input_size} --input_h ${input_size} --name ${dataset}_UKAN --data_dir [YOUR_DATA_DIR]
```
For example, train U-KAN with the resolution of 256x256 with a single GPU on the BUSI dataset in the ```inputs``` dir:
```bash
cd Seg_UKAN
python train.py --arch UKAN --dataset busi --input_w 256 --input_h 256 --name busi_UKAN --data_dir ./inputs
```
Please see Seg_UKAN/scripts.sh for more details.
Note that the resolution of glas is 512x512, differing with other datasets (256x256).
## 🎪Segmentation Model Zoo
We provide all the pre-trained model [checkpoints](https://mycuhk-my.sharepoint.com/:f:/g/personal/1155206760_link_cuhk_edu_hk/Ej6yZBSIrU5Ds9q-gQdhXqwBbpov5_MaWF483uZHm2lccA?e=rmlHMo)
Here is an overview of the released performance&checkpoints. Note that results on a single run and the reported average results in the paper differ.
|Method| Dataset | IoU | F1 | Checkpoints |
|-----|------|-----|-----|-----|
|Seg U-KAN| BUSI | 65.26 | 78.75 | [Link](https://mycuhk-my.sharepoint.com/:f:/g/personal/1155206760_link_cuhk_edu_hk/EjktWkXytkZEgN3EzN2sJKIBfHCeEnJnCnazC68pWCy7kQ?e=4JBLIc)|
|Seg U-KAN| GLAS | 87.51 | 93.33 | [Link](https://mycuhk-my.sharepoint.com/:f:/g/personal/1155206760_link_cuhk_edu_hk/EunQ9KRf6n1AqCJ40FWZF-QB25GMOoF7hoIwU15fefqFbw?e=m7kXwe)|
|Seg U-KAN| CVC-ClinicDB | 85.61 | 92.19 | [Link](https://mycuhk-my.sharepoint.com/:f:/g/personal/1155206760_link_cuhk_edu_hk/Ekhb3PEmwZZMumSG69wPRRQBymYIi0PFNuLJcVNmmK1fjA?e=5XzVSi)|
The parameter ``--no_kan'' denotes the baseline model that is replaced the KAN layers with MLP layers. Please note: it is reasonable to find occasional inconsistencies in performance, and the average results over multiple runs can reveal a more obvious trend.
|Method| Layer Type | IoU | F1 | Checkpoints |
|-----|------|-----|-----|-----|
|Seg U-KAN (--no_kan)| MLP Layer | 63.49 | 77.07 | [Link](https://mycuhk-my.sharepoint.com/:f:/g/personal/1155206760_link_cuhk_edu_hk/EmEH_qokqIFNtP59yU7vY_4Bq4Yc424zuYufwaJuiAGKiw?e=IJ3clx)|
|Seg U-KAN| KAN Layer | 65.26 | 78.75 | [Link](https://mycuhk-my.sharepoint.com/:f:/g/personal/1155206760_link_cuhk_edu_hk/EjktWkXytkZEgN3EzN2sJKIBfHCeEnJnCnazC68pWCy7kQ?e=4JBLIc)|
## 🎇Medical Image Generation with Diffusion U-KAN
Please refer to [Diffusion_UKAN](./Diffusion_UKAN/README.md)
## 🛒TODO List
- [X] Release code for Seg U-KAN.
- [X] Release code for Diffusion U-KAN.
- [X] Upload the pretrained checkpoints.
## 🎈Acknowledgements
Greatly appreciate the tremendous effort for the following projects!
- [CKAN](https://github.com/AntonioTepsich/Convolutional-KANs)
## 📜Citation
If you find this work helpful for your project,please consider citing the following paper:
```
@article{li2024u,
title={U-KAN Makes Strong Backbone for Medical Image Segmentation and Generation},
author={Li, Chenxin and Liu, Xinyu and Li, Wuyang and Wang, Cheng and Liu, Hengyu and Yuan, Yixuan},
journal={arXiv preprint arXiv:2406.02918},
year={2024}
}
```