readme
parents
Showing
nbs/img/dashboard.png
0 → 100644
37.5 KB
nbs/img/forecast.png
0 → 100644
233 KB
nbs/img/forecast_readme.png
0 → 100644
88 KB
nbs/img/logo_nixtlar.png
0 → 100644
41.2 KB
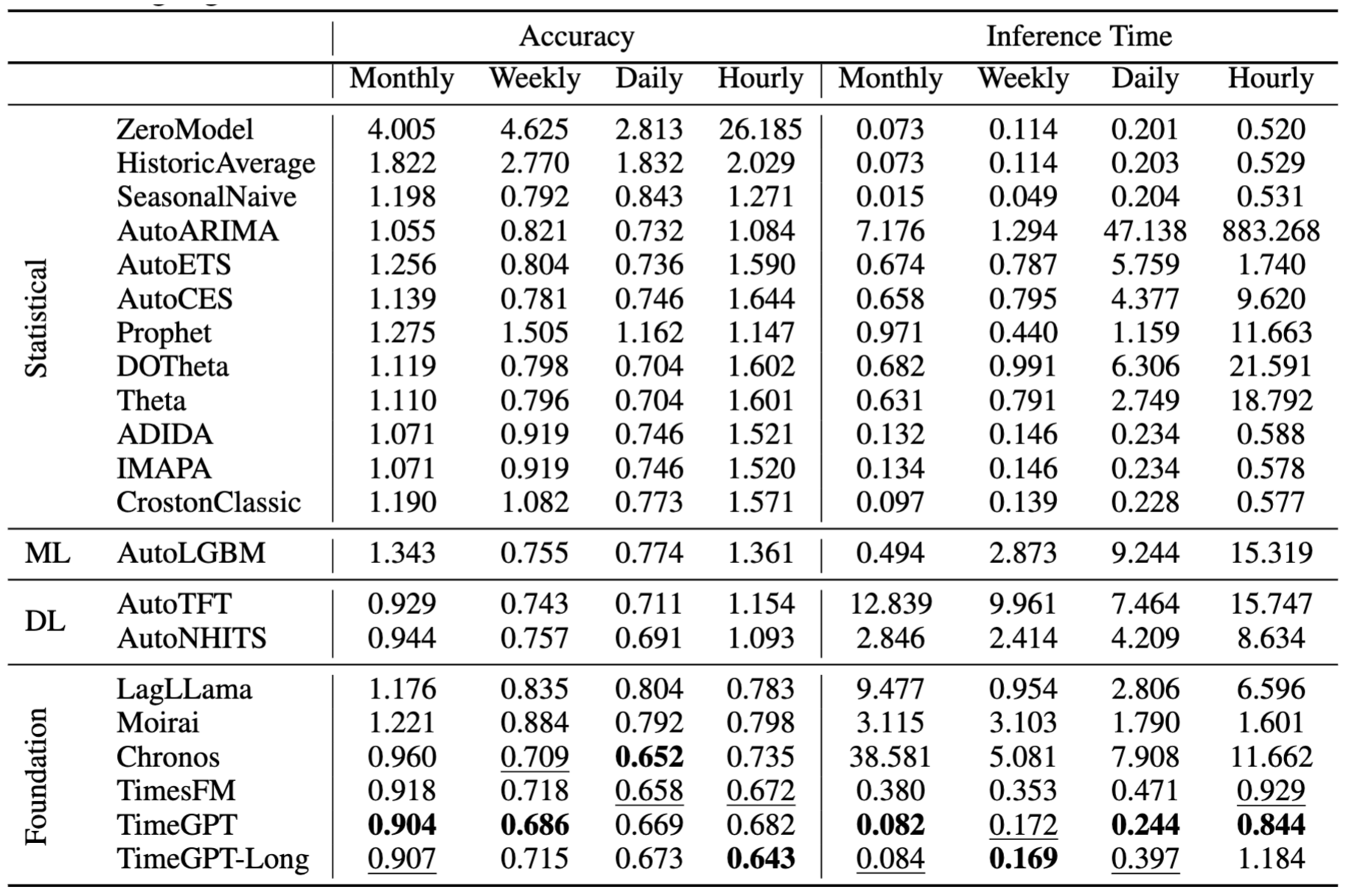
nbs/img/results.jpg
0 → 100644
682 KB
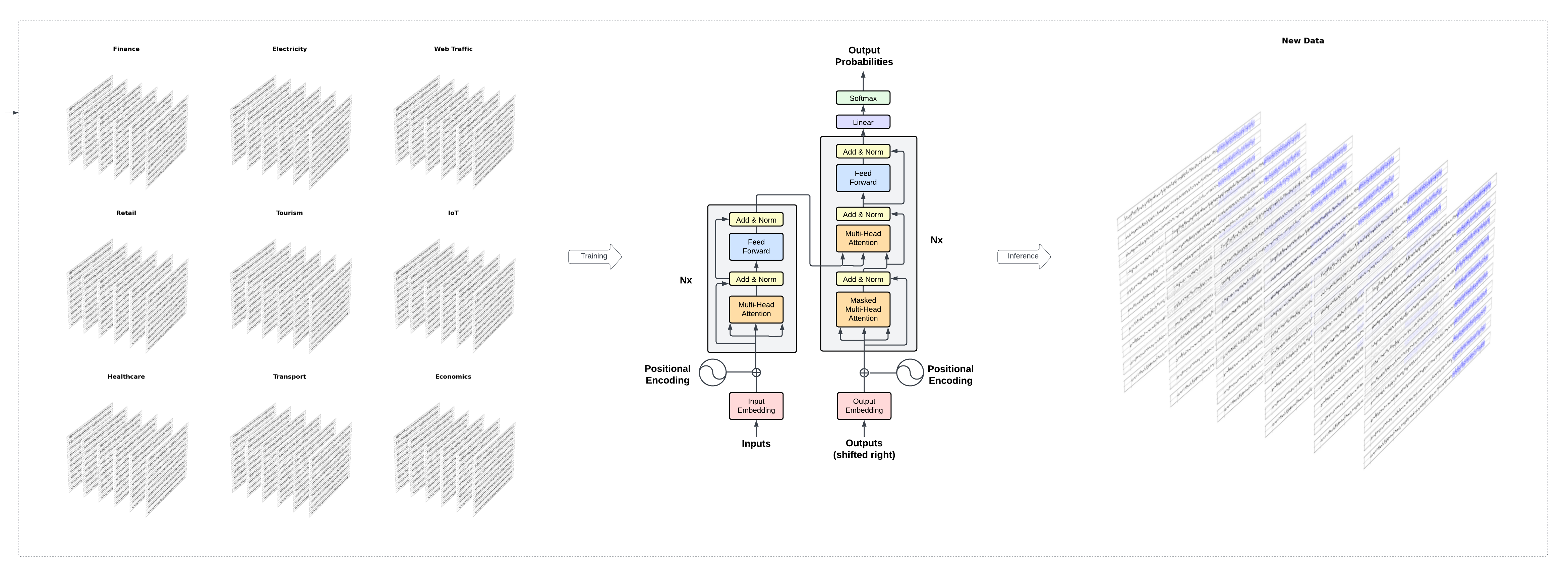
nbs/img/timegpt-arch.png
0 → 100644
2.34 MB
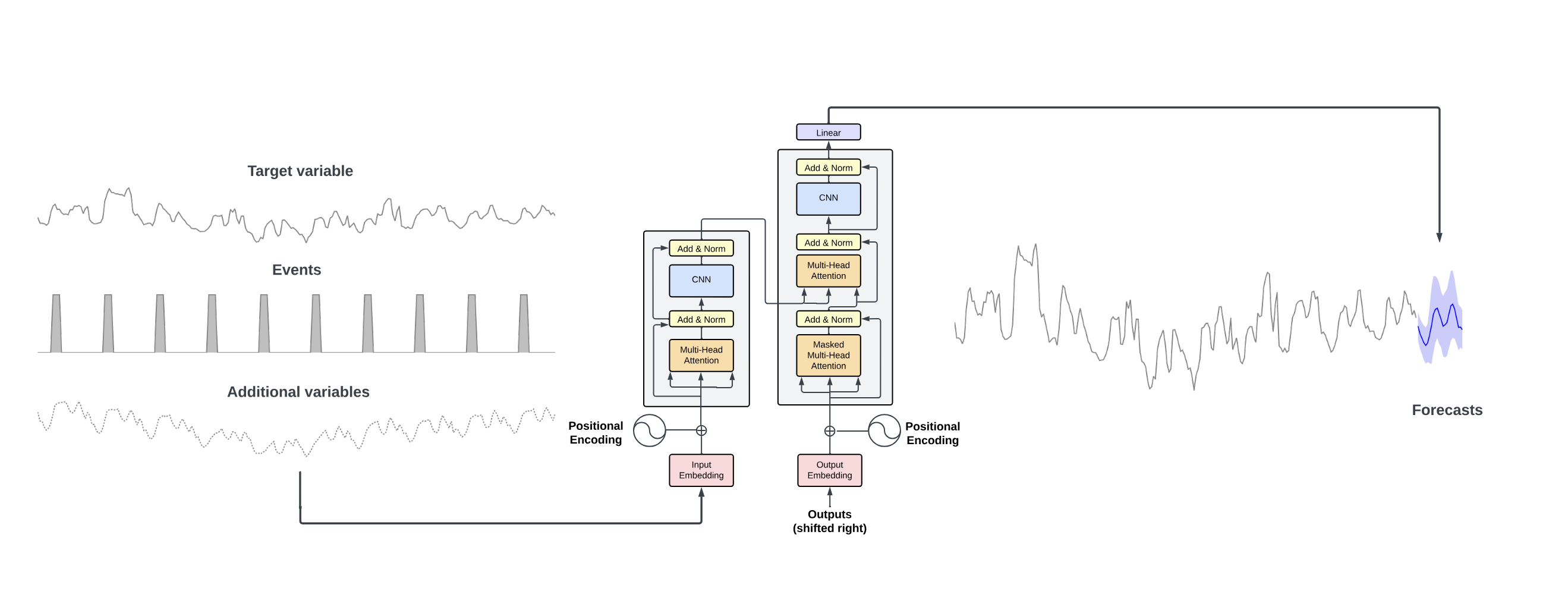
nbs/img/timegpt_archi.png
0 → 100644
233 KB
743 KB
nbs/nbdev.yml
0 → 100644
nbs/sidebar.yml
0 → 100644
nbs/styles.css
0 → 100644
nixtla/__init__.py
0 → 100644
nixtla/date_features.py
0 → 100644
nixtla/nixtla_client.py
0 → 100644
nixtla/py.typed
0 → 100644
nixtla/utils.py
0 → 100644
nixtla_tests/__init__.py
0 → 100644
nixtla_tests/conftest.py
0 → 100644