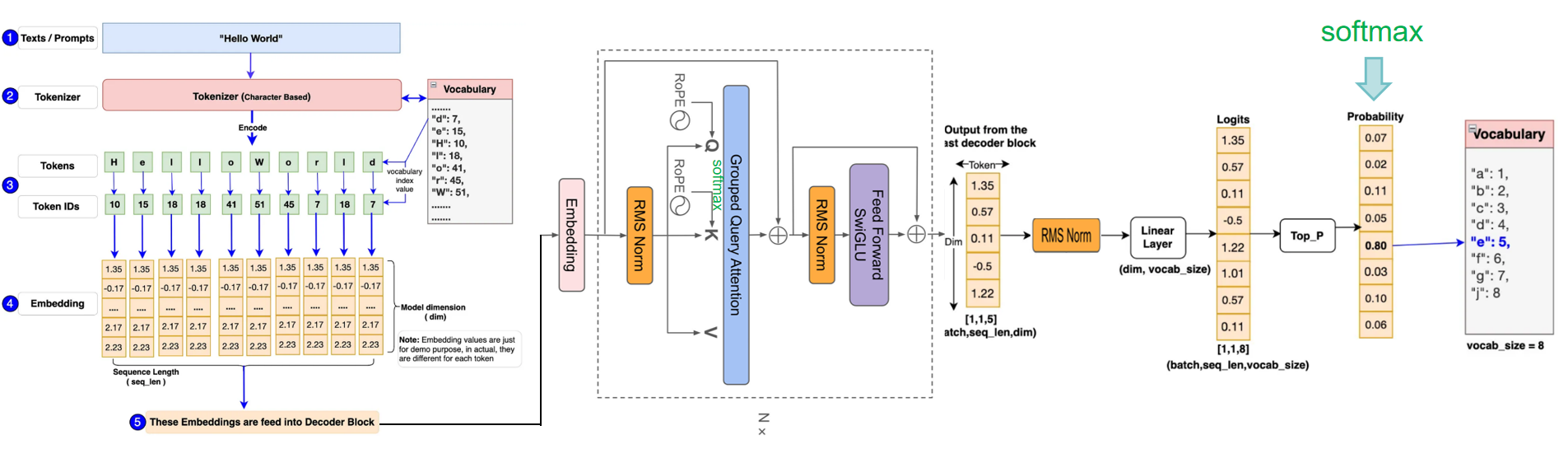
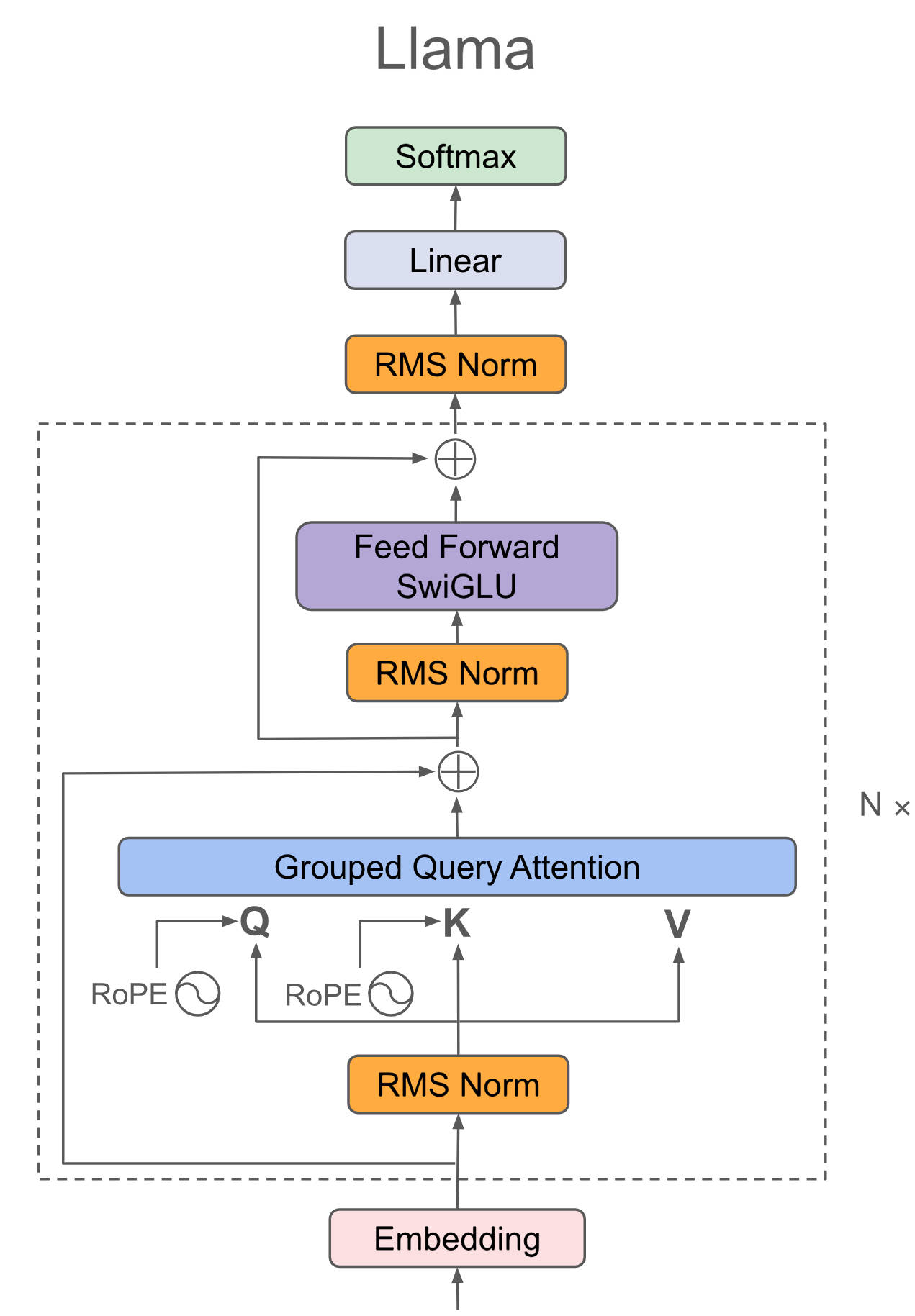
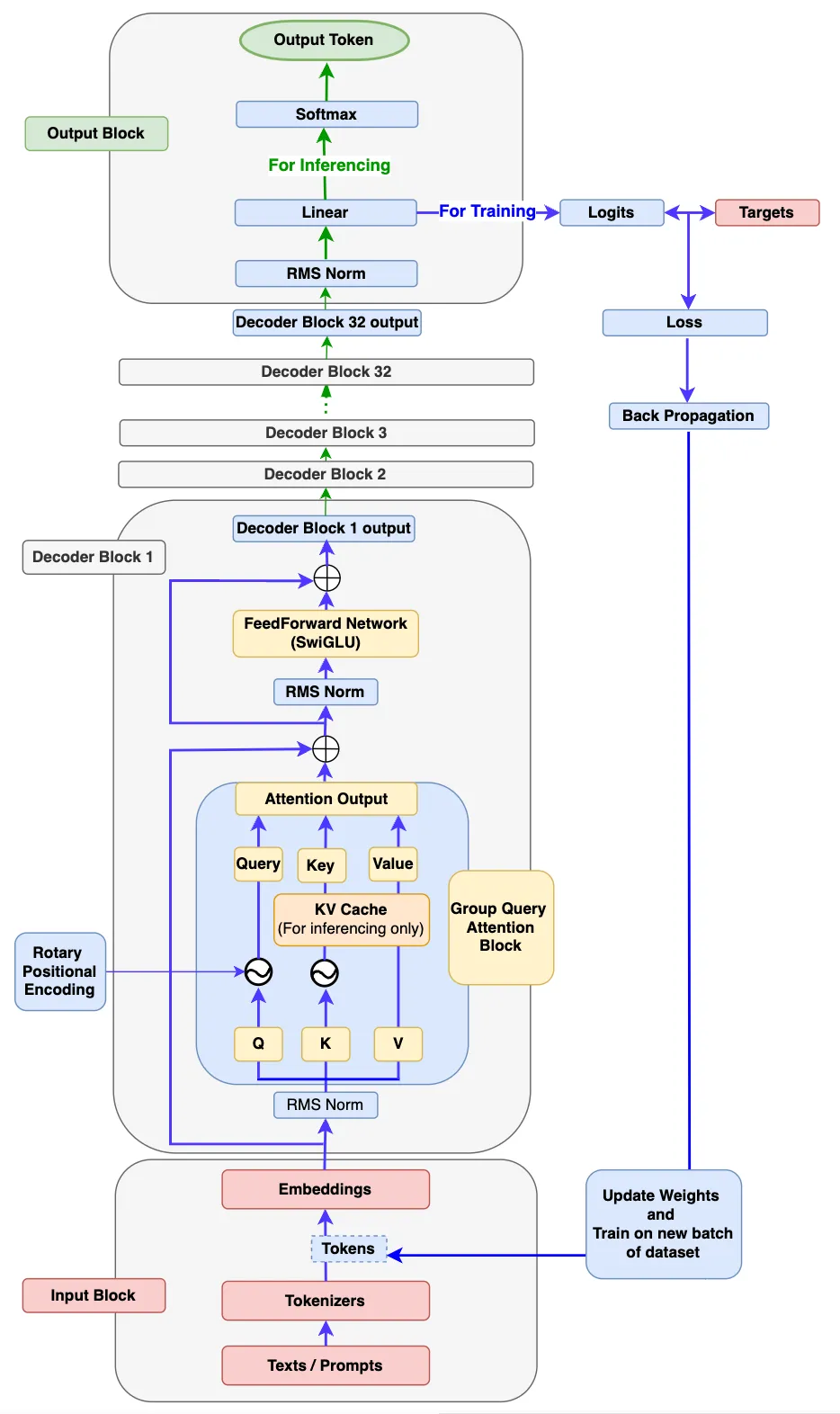
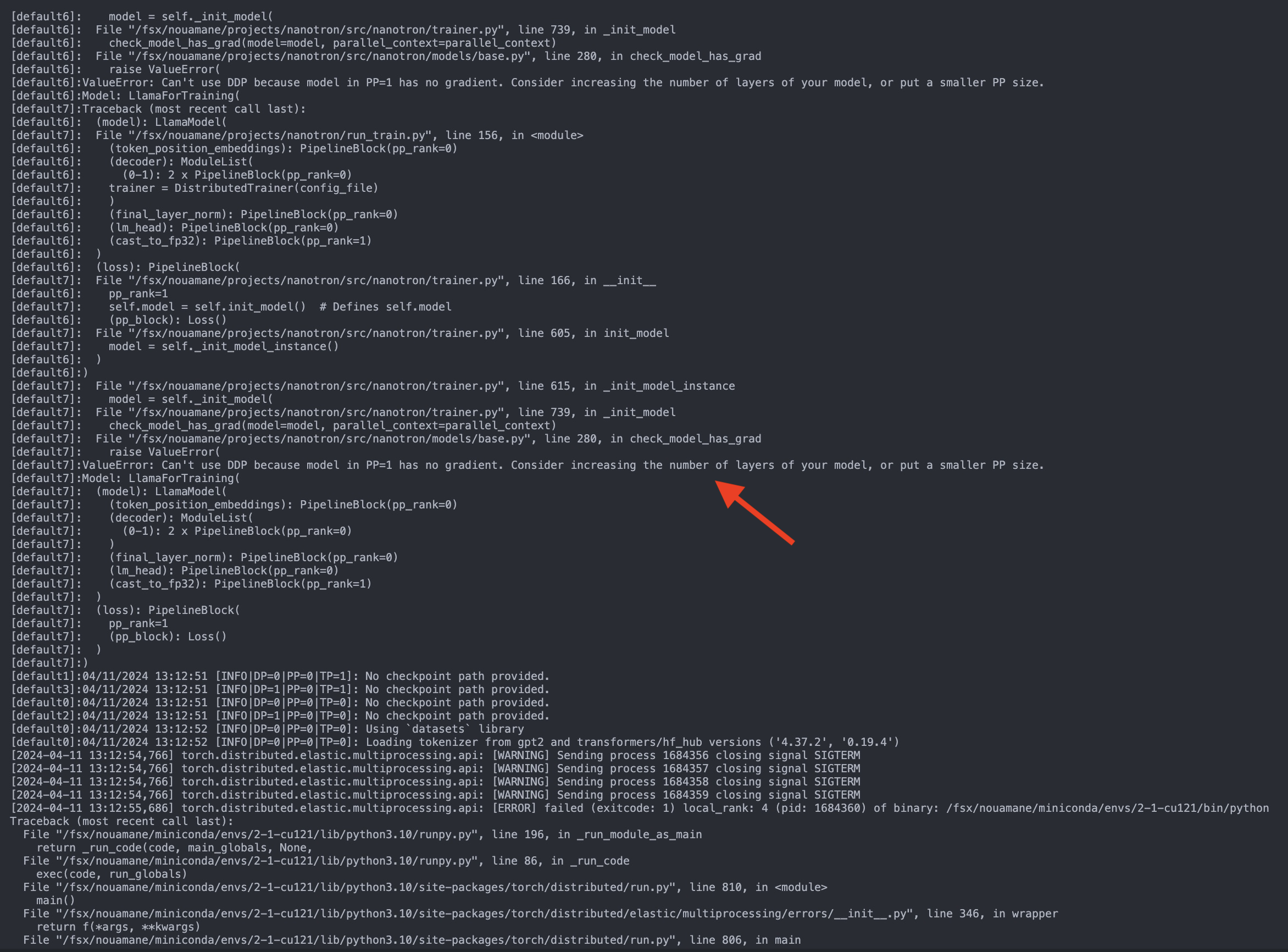
"docs/source/api/pipelines/overview.mdx" did not exist on "a4d5b59f132126c06c1a6b1f266ee44c70440cce"
v1.0.8
parents
Showing
convert_nanotron_to_hf.sh
0 → 100644
doc/algorithm.png
0 → 100644
345 KB
doc/llama3.png
0 → 100644
334 KB
doc/llama3_detail.png
0 → 100644
449 KB
doc/llm.png
0 → 100644
565 KB
docker/Dockerfile
0 → 100644
docker/requirements.txt
0 → 100644
docker_start.sh
0 → 100644
docs/3d_parallelism.md
0 → 100644
docs/debugging.md
0 → 100644
docs/docs.md
0 → 100644
docs/image-2.png
0 → 100644
1.87 MB
docs/image.png
0 → 100644
511 KB
docs/nanoset.md
0 → 100644
examples/bench_llama_7b.py
0 → 100644
examples/config_llama3.yaml
0 → 100644
examples/config_nanoset.yaml
0 → 100644