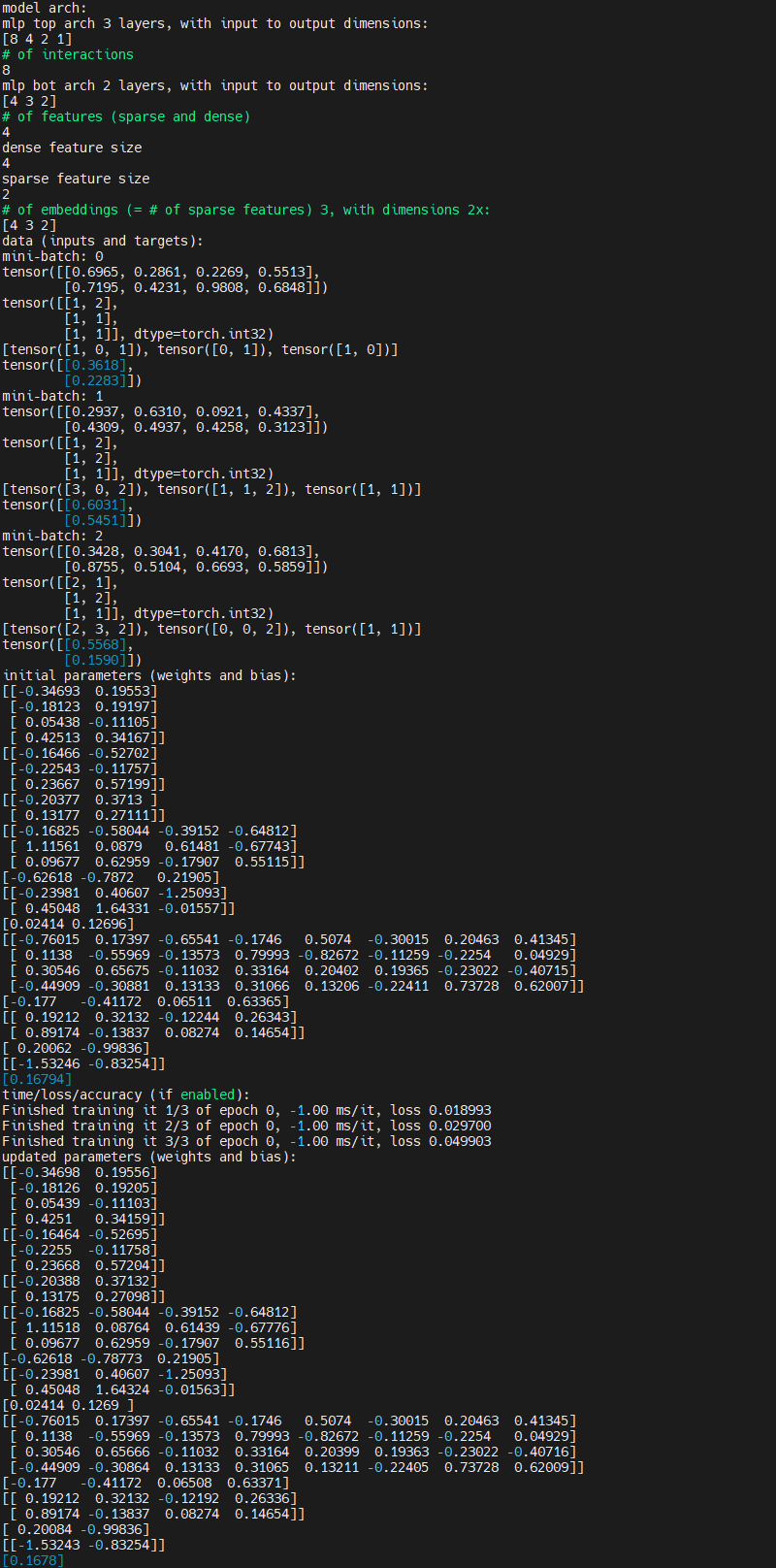
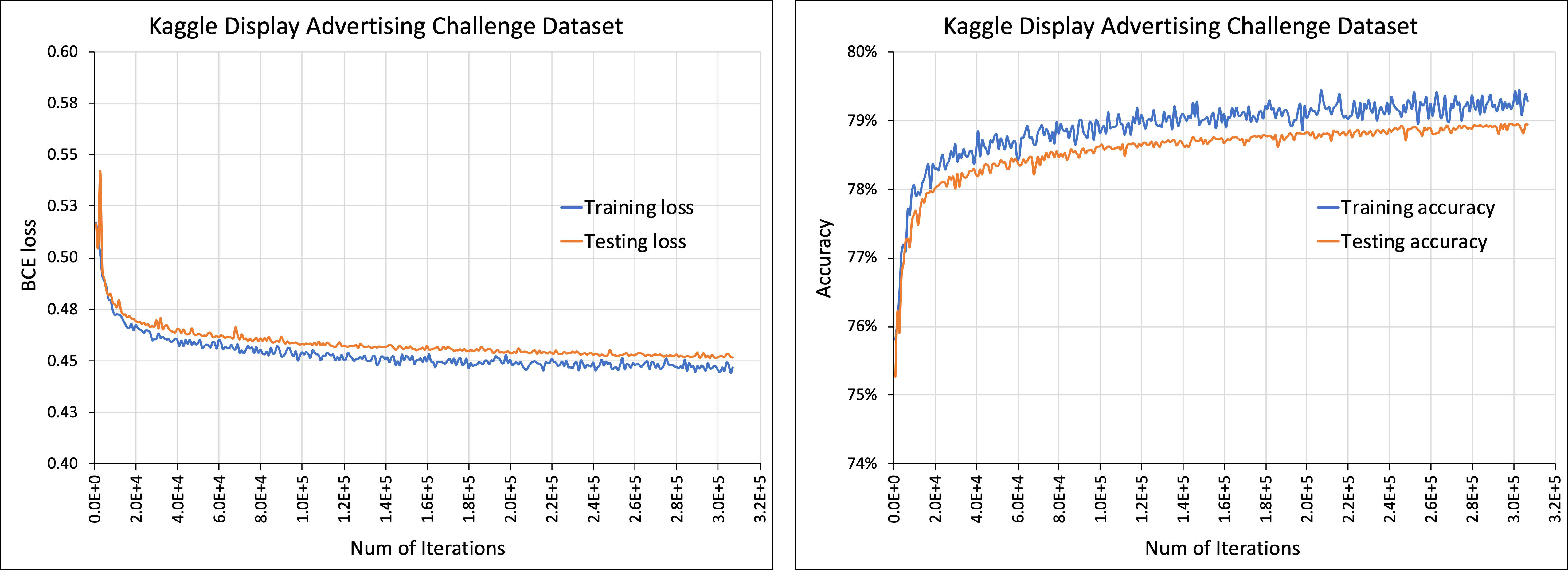
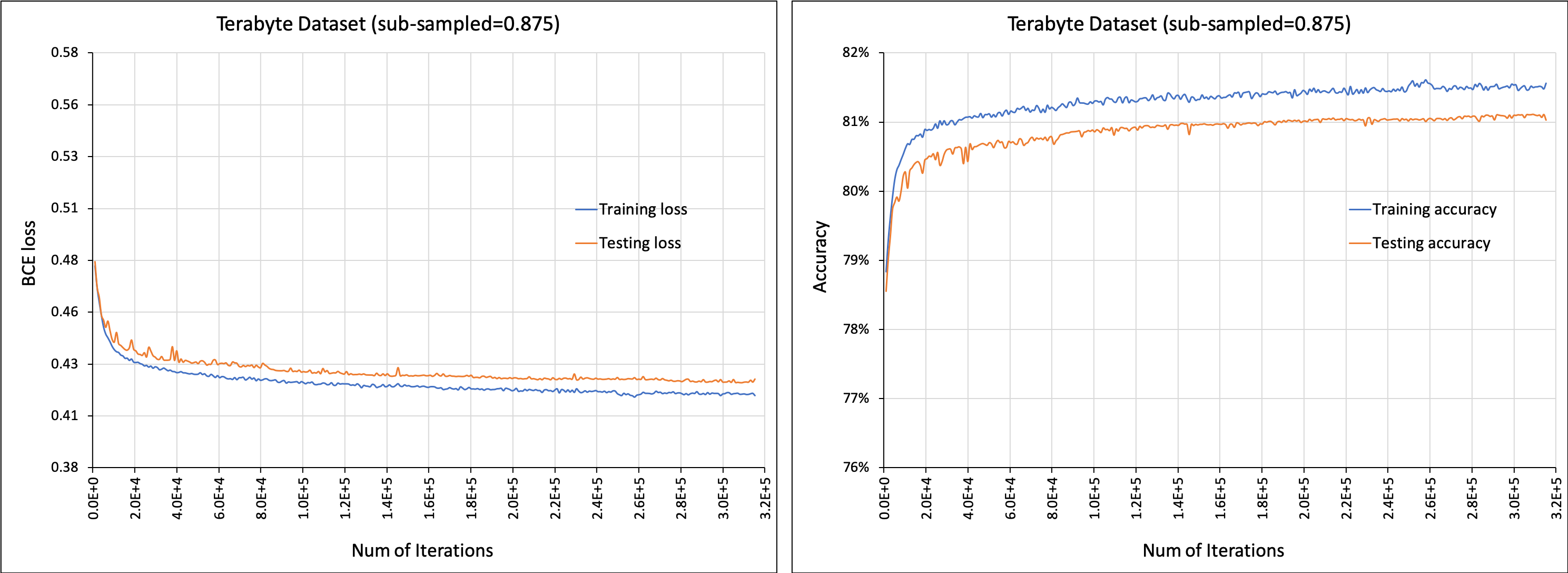
Initial commit
Showing
images/image2.png
0 → 100644
102 KB
images/image3.png
0 → 100644
5.33 KB
535 KB
mlperf_logger.py
0 → 100644
optim/rwsadagrad.py
0 → 100644
requirements.txt
0 → 100644
253 KB
test/dlrm_s_test.sh
0 → 100644
tools/visualize.py
0 → 100644
torchrec_dlrm/Dockerfile
0 → 100644
torchrec_dlrm/README.MD
0 → 100644
torchrec_dlrm/__init__.py
0 → 100644
torchrec_dlrm/dlrm_main.py
0 → 100644