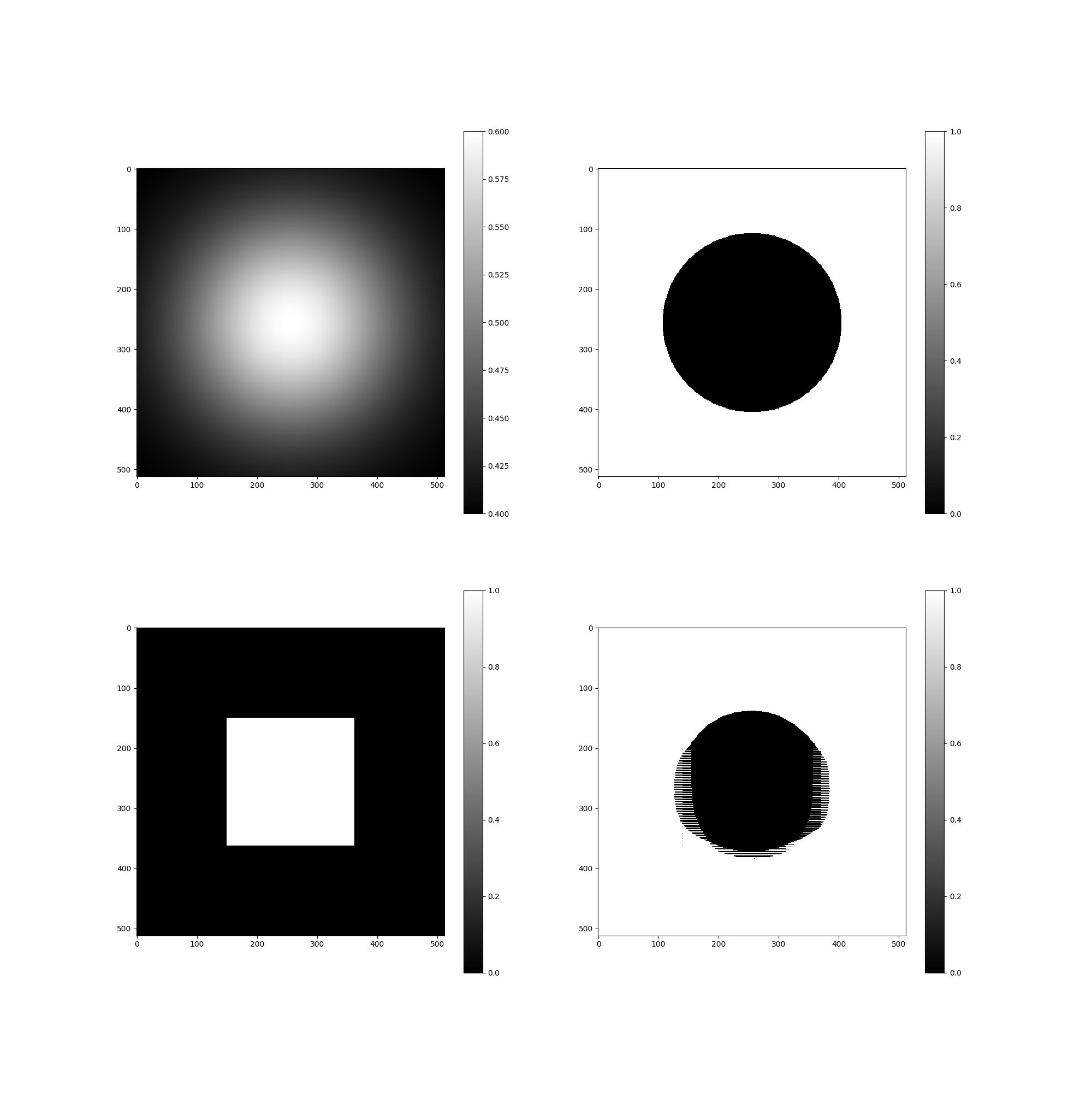
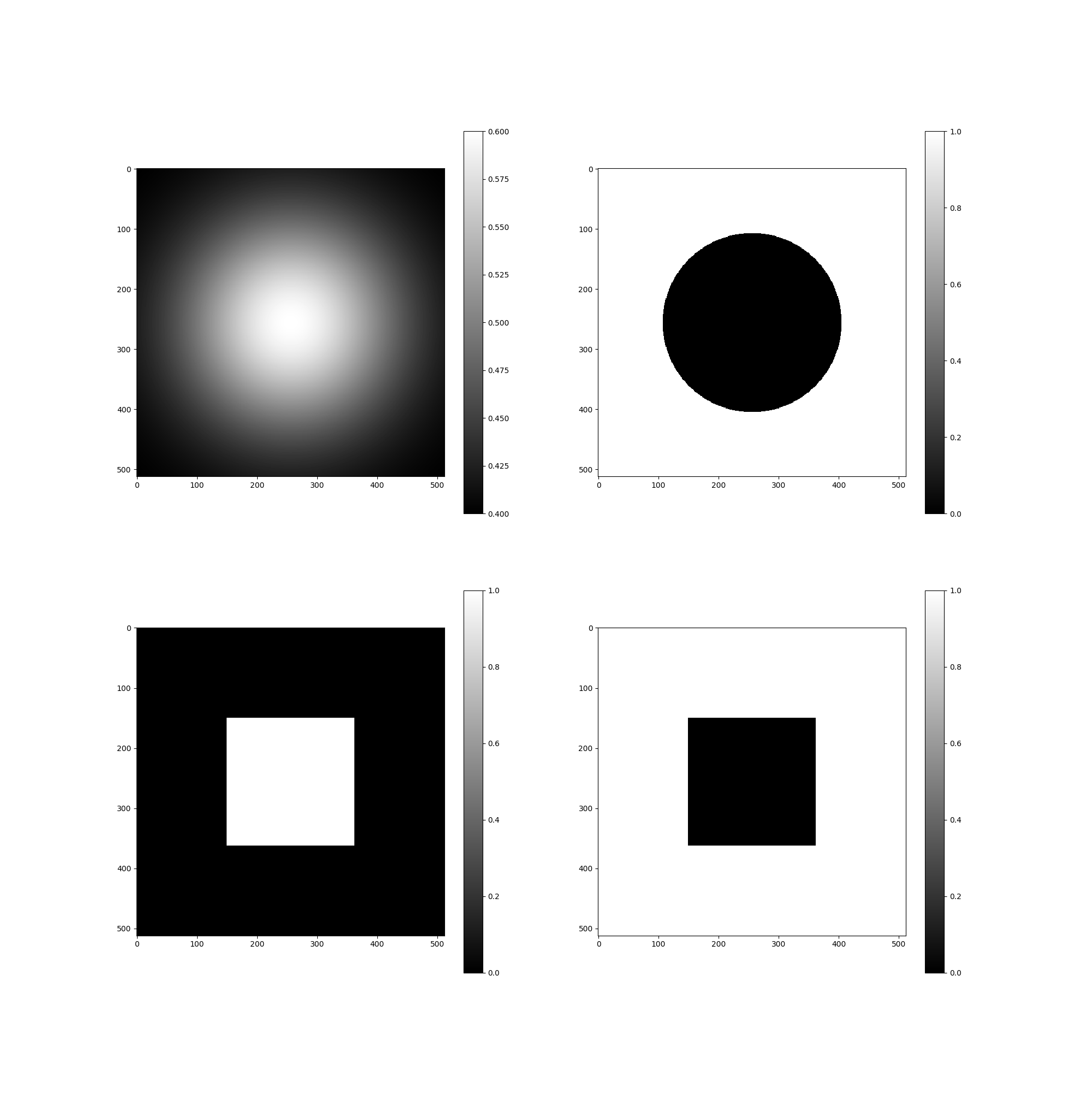
Tests I found that I never commited. Issue #26
Showing
tests/issue26.py
0 → 100644
tests/issue26_problem.png
0 → 100644
128 KB
tests/issue26_solution.png
0 → 100644
126 KB
tests/test_utils.py
0 → 100644