Merge pull request #3029 from liuzhe-lz/v2.0-merge
Merge master into v2.0
Showing
30.4 KB
| W: | H:
| W: | H:


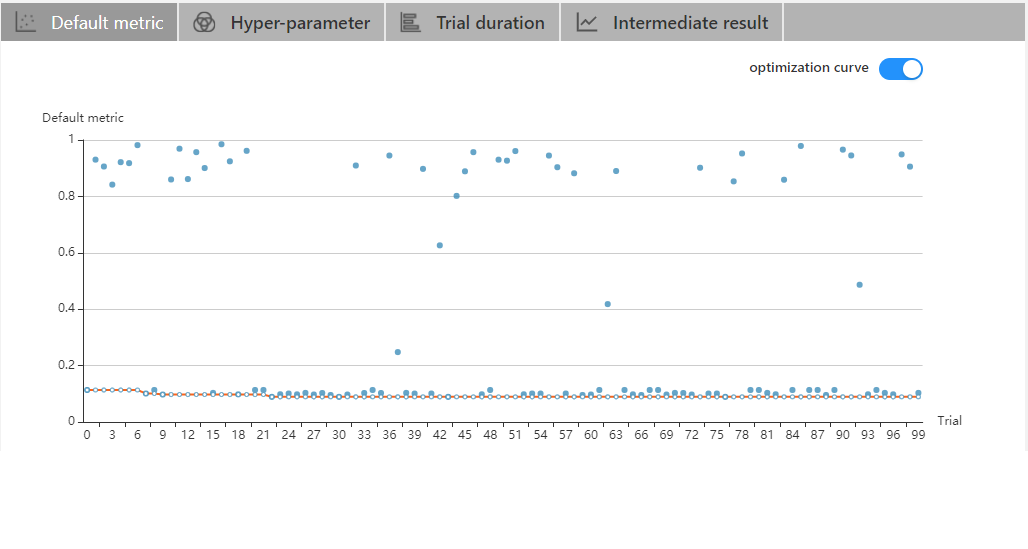
| W: | H:
| W: | H:


Merge master into v2.0
30.4 KB

36.6 KB | W: | H:
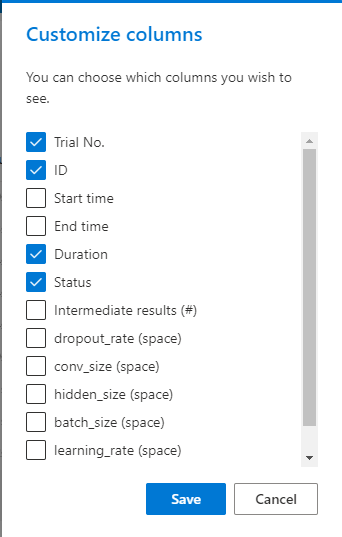
15 KB | W: | H:





32.7 KB | W: | H:
37.3 KB | W: | H:



