Merge pull request #175 from microsoft/master
merge master
Showing
59.6 KB
44.7 KB
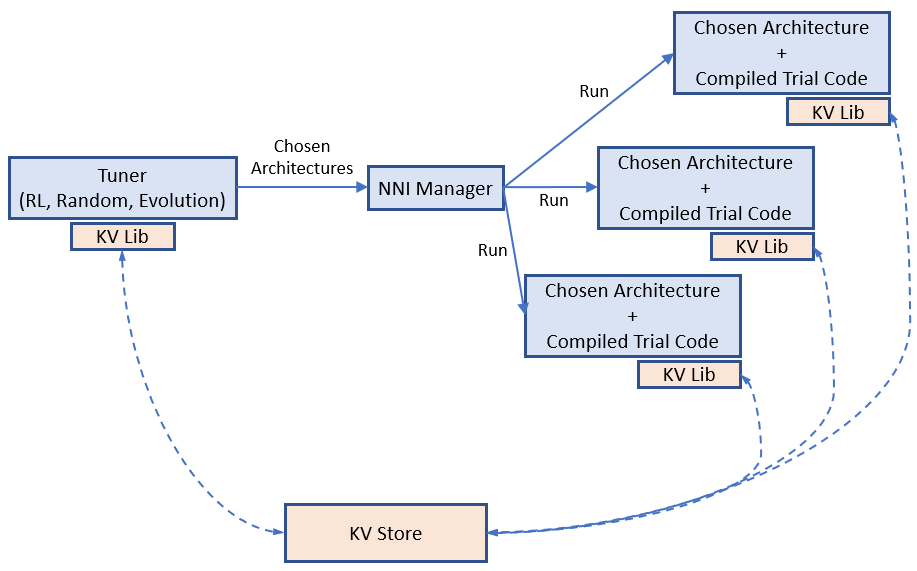
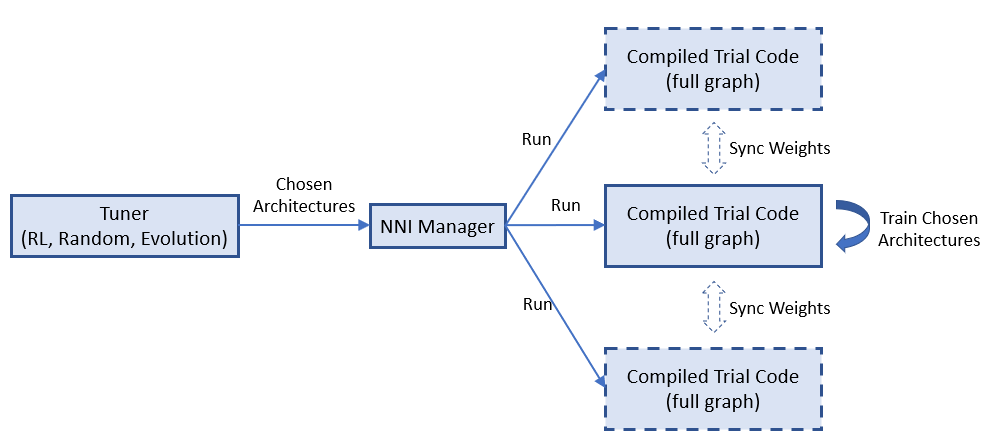
docs/img/example_enas.png
0 → 100644
133 KB
58.6 KB
docs/img/nas_on_nni.png
0 → 100644
40.1 KB
28.9 KB
22.9 KB
merge master
59.6 KB
44.7 KB

133 KB
58.6 KB
40.1 KB
28.9 KB
22.9 KB